- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
I Plate-based Assay for Studying How Fungal Volatile Compounds (VCs) Affect Plant Growth and Development and the Identification of VCs via SPME-GC-MS
(*contributed equally to this work) Published: Vol 9, Iss 4, Feb 20, 2019 DOI: 10.21769/BioProtoc.3166 Views: 8315
Reviewed by: Zhaohui LiuDemosthenis ChronisTimo A Lehti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Ethylene Production in Leaf and Bud Tissue of the Subtropical Tree Crop Litchi (Litchi chinensis Sonn.) Using Gas Chromatography and Flame Ionization Detection
Regina B. Cronje and Arnoldus J. Jonker
Mar 20, 2023 1548 Views

Bi-directional Dual-flow-RootChip for Physiological Analysis of Plant Primary Roots Under Asymmetric Perfusion of Stress Treatments
Claudia Allan [...] Claudia-Nicole Meisrimler
Aug 5, 2023 1945 Views

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1886 Views
Abstract
Biogenic volatile compounds (VCs) mediate various types of crucial intra- and inter-species interactions in plants, animals, and microorganisms owing to their ability to travel through air, liquid, and porous soils. To study how VCs produced by Verticillium dahliae, a soilborne fungal pathogen, affect plant growth and development, we slightly modified a method previously used to study the effect of bacterial VCs on plant growth. The method involves culturing microbial cells and plants in I plate to allow only VC-mediated interaction. The modified protocol is simple to set up and produces reproducible results, facilitating studies on this poorly explored form of plant-fungal interactions. We also optimized conditions for extracting and identifying fungal VCs using solid phase microextraction (SPME) coupled to gas chromatography-mass spectrometry (GC-MS).
Keywords: Arabidopsis thalianaBackground
Volatile compounds (VCs) have been shown or suggested to play varied and crucial roles in mediating organismal interactions within and across kingdoms. Plants rely on VCs to attract pollinators, seed dispersers, and parasitoids (Baldwin, 2010; Herrmann, 2010). Animals have evolved sophisticated olfactory systems to detect and respond to foods, threats, and mates through volatile cues (Buck, 2004). Similarly, microbial VCs seem to perform diverse functions such as suppressing competitors, regulating their population density, and controlling morphological transitions (Bailly and Weisskopf, 2012; Bennett et al., 2012; Bitas et al., 2013). Roles of microbial VCs in plant growth, development, and stress response have been investigated using several experimental setups that physically separate microbial cells from plants so that only VC-mediated interaction can occur (Ryu et al., 2003; Kai and Piechulla, 2009; Xie et al., 2009; Hung et al., 2013; Vaishnav et al., 2015). Among them, a method employing bipartite Petri plate, also called I plate, has been most frequently used. We adopted I plate to study the effect of VCs produced by soilborne fungal pathogens on plant growth, development, and responses to biotic and abiotic stresses (Bitas et al., 2015; Li and Kang, 2018; Li et al., 2018b). In addition, we optimized a scheme for VC extraction and analysis to help identify fungal VCs that are responsible for modulating plant growth and development (Li et al., 2018b).
Here, we provide a detailed protocol for setting up an I plate assay used for evaluating the effect of VCs produced by Verticillium dahliae, a devastating soilborne fungal pathogen that infects hundreds of plant species, on Arabidopsis thaliana. This protocol enables rapid and straightforward determination of if and how fungal VCs affect plants. We also describe a protocol for capturing VCs through solid phase microextraction (SPME) and analyzing extracted VCs via gas chromatography-mass spectrometry (GC-MS). In combination, these protocols will help explore how VCs produced by diverse fungi affect plants and can also be applied to study VC-mediated interactions between microbes.
Materials and Reagents
- Surgical blades #10 and #11
- Parafilm (Bemis, catalog number: PM-99)
- Paper towel
- 10 µl and 1,000 µl micropipette tips
- 100 x 15 mm I plate (VWR, catalog number: 25384-310)
- 100 x 100 mm square plate (VWR, catalog number: 10799-140)
- 100 x 15 mm (VWR, catalog number: 25384-302) and 60 x 15 mm (VWR, catalog number: 25384-092) Petri plates
- 1.7 ml microcentrifuge tube (VWR, catalog number: 87003-294)
- 25 ml serological pipette (VWR, catalog number: 89130-900)
- Filter unit with 0.2 µM cellulose membrane (Nalgene, catalog number: 121-0020)
- 1.5 ml sample vial (Shimadzu, catalog number: 221-34274-91), white cap with septum
- V. dahliae strains PD322 and PD413 (conidial suspension in 20% glycerol and stored at -80 °C)
- A. thaliana ecotype Col-0 seeds (Lehle Seed Co.)
- Sterile MilliQ water
- Murashige and Skoog (MS) basal medium (Sigma-Aldrich, catalog number: M0404-10L)
- Sucrose (Alfa Aesar, catalog number: A15583)
- Granulated agar (Difco, catalog number: 214530)
- Potato dextrose agar (PDA) (Difco, catalog number: 213400)
- 200 proof ethanol (KOPTEC, catalog number: 64-17-5)
- 6% sodium hypochlorite (CLOROX)
- n-Hexane (EMSURE, catalog number: 1043744000)
- C7-C30 saturated alkanes (Sigma-Aldrich, catalog number: 49451-U)
- 99.99% pure helium gas
- 0.5x PDA medium (see Recipes)
- MS agar medium (see Recipes)
Equipment
- Pyrex glass bottle
- 10 µl and 1,000 µl micropipettes
- Scalpel
- Forceps
- Cork borer (5 mm in diameter)
- SPME fiber holder (Supelco, catalog number: 57330-U)
- 15 ml clear glass vial (Supelco, catalog number: 27159), screw cap with PTFE/silicone septum
- SPME fiber assembly with 50/30 µm DVB/CAR/PDMS fiber coating (Supelco, catalog number: 57328-U)
- Electric pipette controller (Drummond Scientific Co., catalog number: 4-000-110-TC)
- Vortex (VWR, model: Genie 2)
- Table-top centrifuge (Eppendorf, model: 5417C)
- Table-top shaker (VWR, catalog number: 57018-754)
- Analytical balance (Mettler Toledo, model: AE-100)
- Dissecting microscope (Zeiss, model: Stemi 2000-C)
- Incubator (Sheldon Manufacturing, model: 1510E)
- Plant growth chamber (Conviron, model: CMP5090)
- Flexible-arm electrode holder (Mettler Toledo, catalog number: 30266628)
- GC-MS system (Shimadzu, model: GCMS-QP2010 ultra) equipped with AOC-20i auto injector (Shimadzu, catalog number: 221-72315-48)
- Rtx-Wax capillary (60 m, 0.25 mm ID and 0.25 μm df) column (Restek, catalog number: 12426)
- 4 °C refrigerator
- Autoclave
Software
- ImageJ (version 1.52a)
- GC-MS Solution (Shimadzu, version 2.72), a package supporting GC-MS real-time and post-run analyses
- National Institute of Standards and Technology (NIST) Mass spectral library (Shimadzu, version 11)
Procedure
- I plate assay (Figure 1)
- Use a sterile 10 µl pipette tip to streak V. dahliae stock on 0.5x PDA (Recipe 1) plate in a zigzag pattern and incubate at 22 °C for 10 days.
- Surface sterilize A. thaliana seeds as follows:
- Add 1 ml 95% ethanol to a 1.7 ml microcentrifuge tube containing seeds, vortex, and incubate for 1 min.
- After removing ethanol, wash once with sterile MilliQ water and discard the water.
- Add 1 ml 6% sodium hypochlorite solution, vortex, and incubate for 15 min by shaking at 100 rpm.
- Wash twice with sterile MilliQ water after removing the sodium hypochlorite solution.
- Incubate seeds in 1 ml sterile MilliQ water at 4 °C in darkness for 3 days.
- Prepare square plates with MS agar (Recipe 2) and slice the medium into 10 x 10 mm pieces using a sterilized scalpel with blade #10.
- Hold one seed at the tip of a 10 µl micropipette using suction and then release the seed onto each agar piece. Seal the plate with two layers of Parafilm and place in a plant growth chamber set at 22 °C, 12 h light (4,500 lux, 60 μmol photons m-2 s-1), and 60% relative humidity for 7 days.
- Prepare I plate by adding 8 ml MS agar to one compartment and 8 ml 0.5x PDA to the other compartment.
- Transfer five A. thaliana seedlings (similar in size and growth stage) along with attached agar piece to the MS side of I plate using a sterilized scalpel with blade #10.
- Use a heat-sterilized cork borer to generate culture plugs along the actively growing margin of V. dahliae culture and place one plug (upside down) to the far end of the PDA side of I plate using a sterilized scalpel with blade #11.
- Seal the inoculated I plate with two layers of Parafilm and place in a plant growth chamber for a designated amount of time for each experiment (see Li et al., 2018b for specific examples).
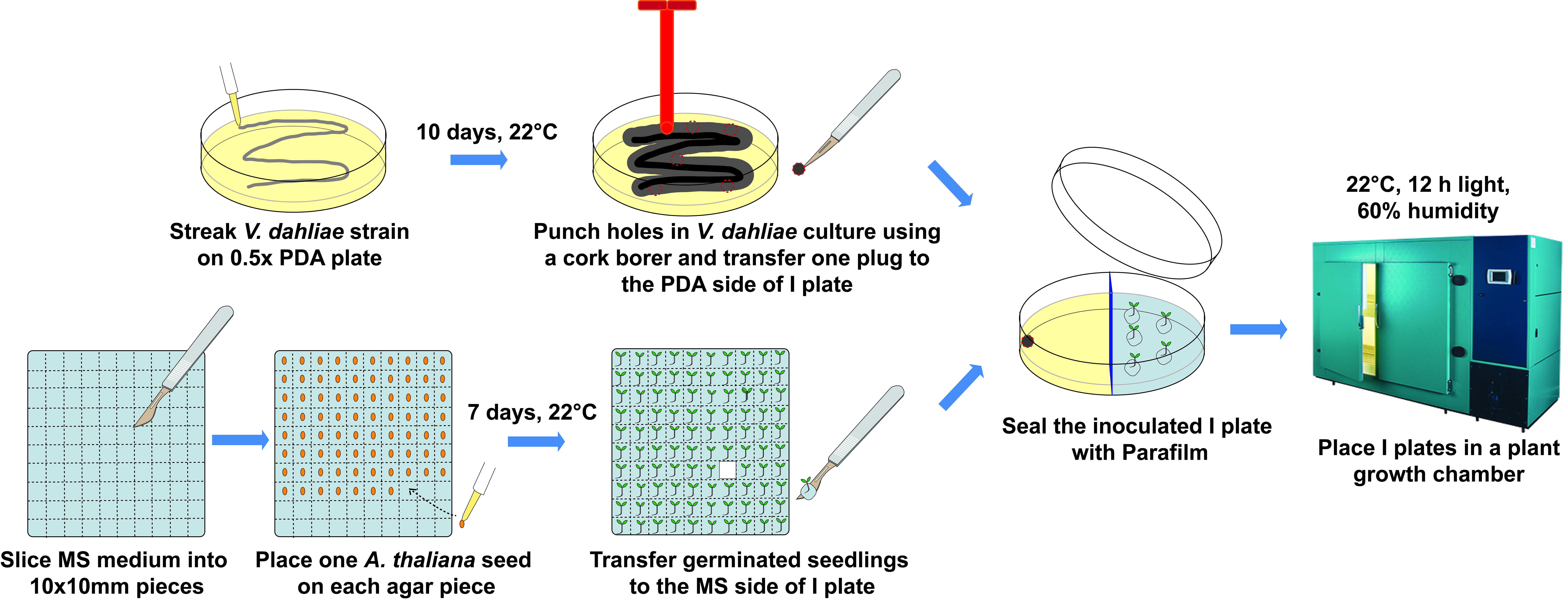
Figure 1. Workflow of the I plate-based assay - Extraction of VCs produced by V. dahliae (Figure 2)
- Inoculate a plug of V. dahliae culture on 8 ml PDA slant in a 15 ml clear glass vial. Seal the vial with Parafilm.
- Incubate at 22 °C until the culture fully covers the surface of PDA slant, which typically takes 8 days.
- Replace Parafilm with a screw cap containing PTFE/silicone septum and incubate for one day.
- Condition the SPME fiber before VC extraction by placing the SPME needle into the GC injection port set at 230 °C for 1 h.
- Insert the conditioned SPME fiber in the injection port and starting the GC temperature program shown in Table 1. Desorb the fiber for 5 min in the injection port. Retract the fiber and remove the needle after 5 min and check resulting chromatograms when the program is completed. The intensity of background peaks (= blank sample) should be very low. Otherwise, repeat the desorption of the fiber.
- Extract VCs for 1 h by leaving the SPME fiber in the headspace of sampling vial. Use a flexible-arm electrode holder to lock the position of the SPME fiber holder so that the insertion depth of fiber is uniform for all extractions.
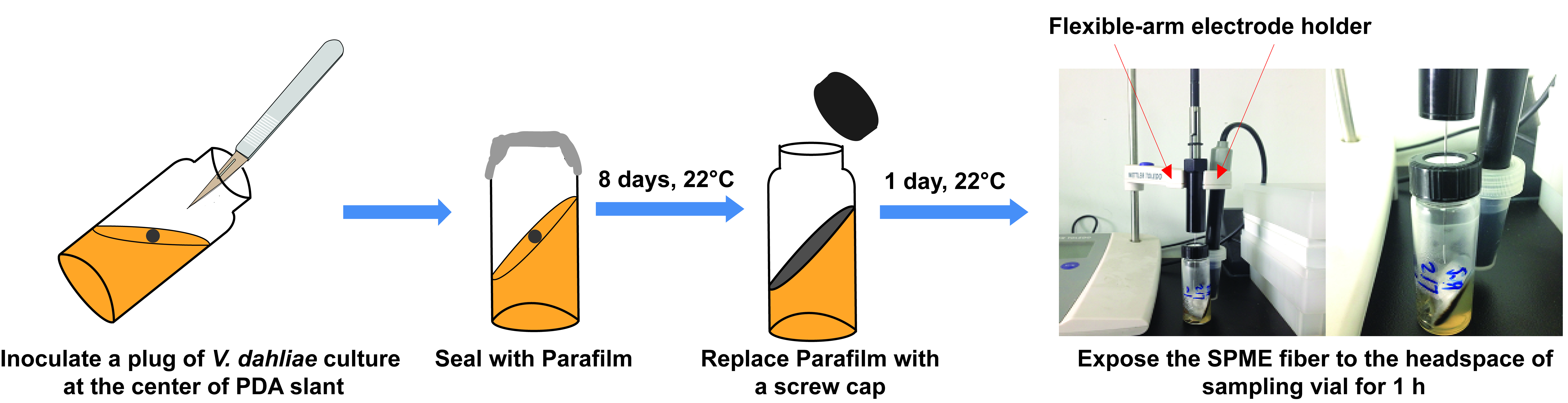
Figure 2. Workflow of volatile compound extraction
Table 1. Conditions for gas chromatography-mass spectrometry analysis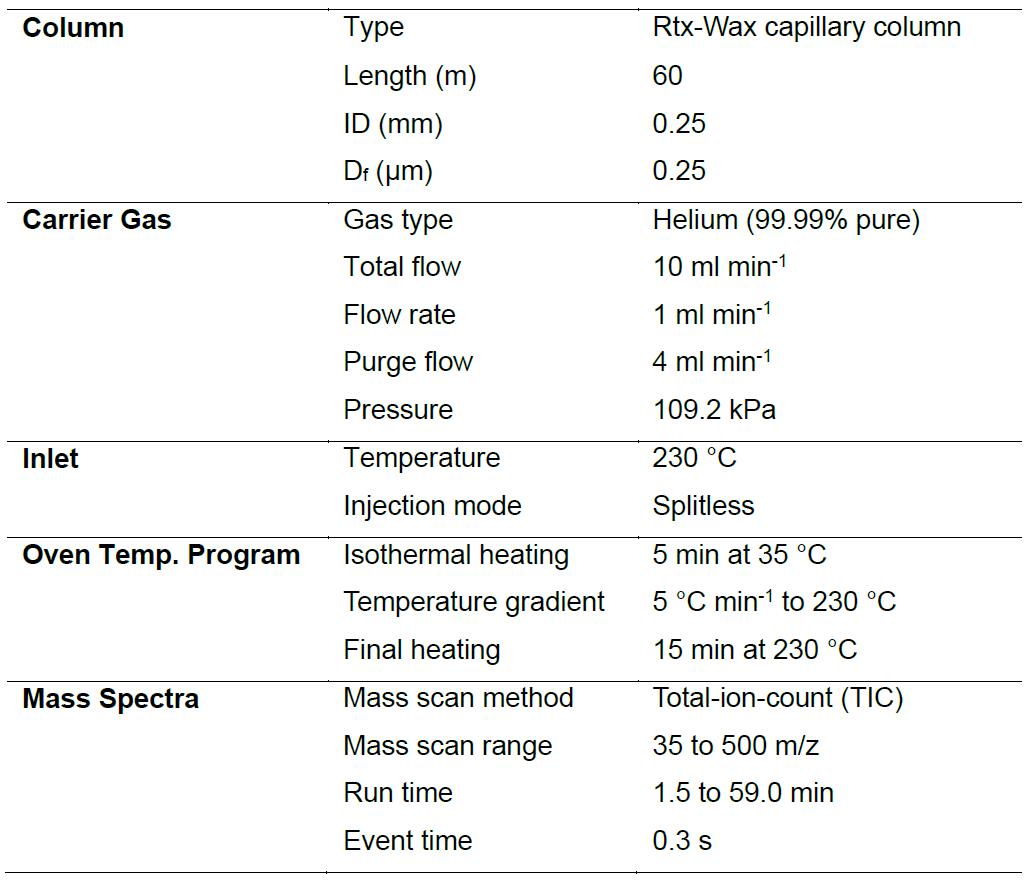
- GC-MS analysis of extracted VCs (Figure 3)
- Analyze the extracted VCs using a GC-MS system (manual injection mode, Figure 3A) by following the conditions described in Table 1. Insert the SPME needle into the GC injection port immediately after retracting it from each sampling vial. Desorb VCs bound to the SPME fiber for 5 min. An Rtx-Wax capillary column is used as it exhibits better retention and separation of polar VCs.
- Place the fiber in the GC injection port for 20 min for desorption before extracting another sample of VCs.
- Analyze C7-C30 saturated alkane standards using the automatic sample injection mode (Figure 3B). Connect the AOC-20i auto injector to the GC injection port. Place a 1.5 ml sample vial containing 0.5 ml solution of standards (1:50 dilution with n-hexane) on the rack of the auto injector. Inject 1 μl solution into the GC-MS system and analyze using the same column and conditions (Table 1).
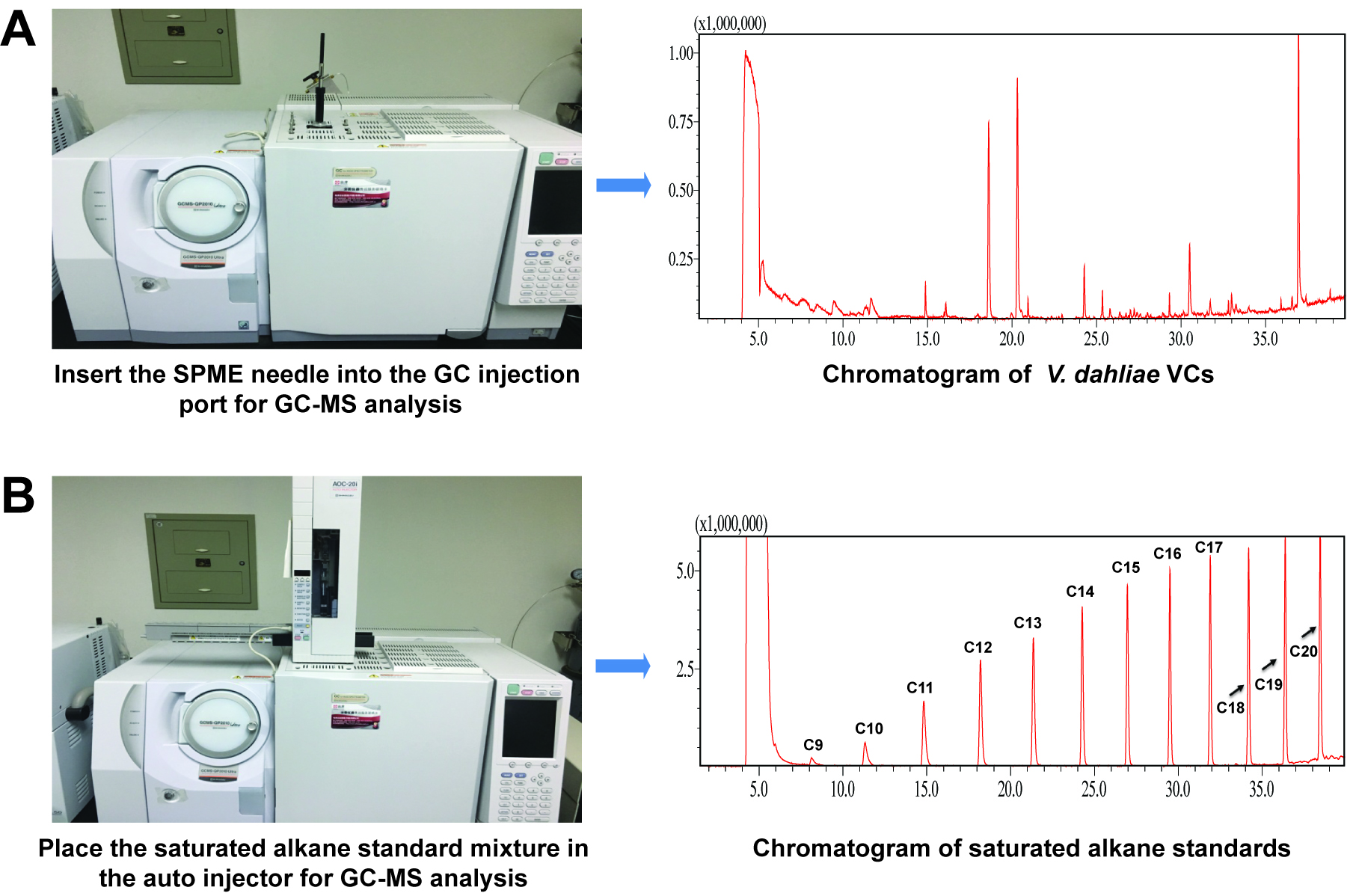
Figure 3. Gas chromatography-mass spectrometry analysis. A. Extracted volatile compounds (VCs) from V. dahliae were analyzed via the manual injection mode. B. Alkane standards were analyzed using the automatic sample injection mode.
Data analysis
- Assessment of root growth and development after individual VC treatments
- After co-cultivation, use forceps to gently pull A. thaliana seedlings from the medium without damaging their roots.
- Remove excess moisture on the roots using a paper towel.
- Detaching roots from the shoot using forceps, weigh the roots immediately using an analytical balance.
- Mount the roots on a flat surface (e.g., 100 x 15 mm Petri plate) and add 3 ml water on them.
- Gently spread the roots using forceps so that the primary and lateral roots do not overlap.
- Use the bottom of 60 x 15 mm Petri plate to flatten the roots (Figure 4A) for subsequent measurements:
- Count the number of lateral roots, including all branches, under a dissecting microscope.
- Take pictures of the roots and import them to ImageJ. Follow the instruction in ImageJ to measure the primary root length (Figure 4B).
- Calculate the lateral root density of each sample by dividing the number of lateral roots by the primary root length.
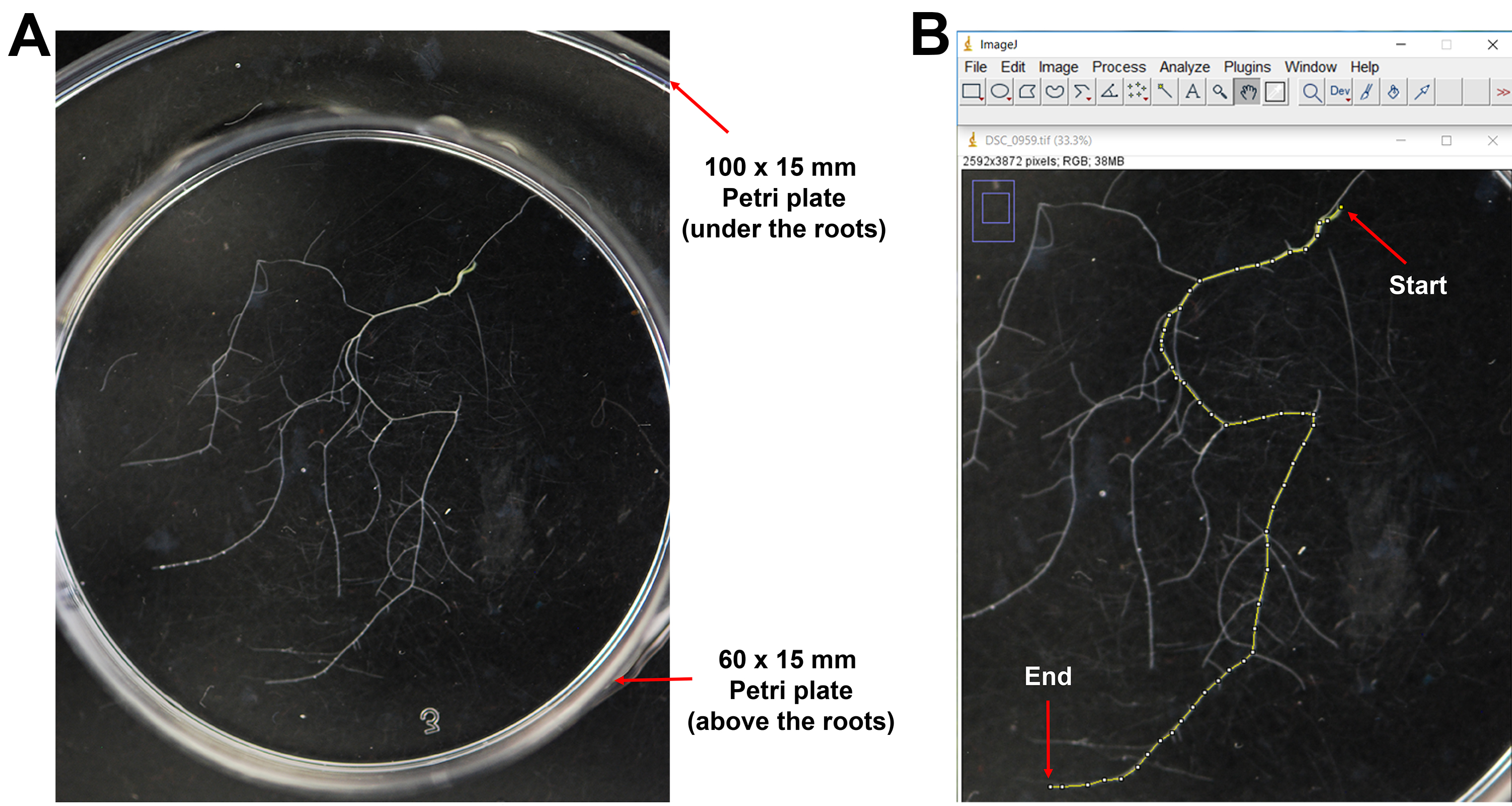
Figure 4. Analysis of A. thaliana roots after treating with volatile compounds produced by V. dahliae. A. Flattened A. thaliana roots by pressing with a 60 x 15 mm Petri plate. B. Snapshot of ImageJ data used to measure the length of the primary root (indicated by the yellow line) after VC treatment. The start and end of the primary root were indicated using red arrows.
- Identification of individual compounds
- Peak finding, peak integration, and retention time correction were performed using the post-run analysis software in the GC-MS Solution package.
- The putative identity of each compound (peak) was determined using the following methods.
- Compare resulting mass spectral profiles with reference data archived in the NIST mass spectral library (version 11). The top hit(s) with match factors ≥ 90% were put on a “positive list” of tentatively identified compounds.
- Calculate the retention index (RI) of each compound using the following equation (for temperature programmed chromatography) (Kováts, 1958).
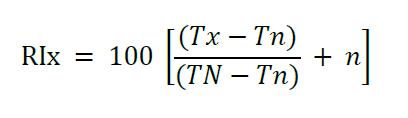
where,
x = Unknown compound in the sample
n = The number of carbons in the alkane preceding the unknown compound
N = The number of carbons in the alkane following the unknown compound
Tx = The retention time of the unknown compound
Tn = The retention time of the preceding alkane
TN = The retention time of the following alkane
The experimentally obtained RI of each peak was compared to those in the NIST Chemistry WebBook (https://webbook.nist.gov, using polar columns). For positive confirmation of identity, a maximum relative deviation of ± 2% from published values was used (Stoppacher et al., 2010).
Notes
- The I plate assay may not be suitable for studying the effect of VCs produced by fungi that grow rapidly, as fungal mycelia may grow over the central divider of I plate and contaminate the MS medium. We removed agar strips from both sides of the divider of I plate to prevent this kind of contamination when we studied fungi that grow faster than V. dahliae (Bitas et al., 2015). However, for fungi like Trichoderma, this measure was not sufficient (Li et al., 2018a).
- Due to the limited space of I plate, only small plants (e.g., A. thaliana and Nicotiana benthamiana) in their early growth stage are suitable for this assay.
- Under the growth conditions used, growth promoting effect on A. thaliana becomes noticeable as early as after 7 days of co-cultivation with V. dahliae (Li et al., 2018b).
- Because A. thaliana starts to initiate inflorescence development after 14 days of co-cultivation, we recommend that the duration of co-cultivation should not exceed 14 days.
- Here, we only described how to analyze the root growth and development after VC exposure. Other traits of VC-exposed plants, such as shoot weight, chlorophyll content, physiological and molecular changes, can also be analyzed (e.g., Zhang et al., 2007 and 2008; Li et al., 2018b).
- Several SPME fibers and GC columns may need to be evaluated to optimize the scheme for VC extraction and analysis. In our research, the DVB/CAR/PDMS 50/30 µm fiber coating extracted the largest number of VCs, and the Rtx-Wax capillary column enabled better separation of extracted VCs than the DB-5 column.
- Analysis of VCs extracted from uninoculated PDA slant is necessary to exclude VCs derived from the medium and the environment.
Recipes
- 0.5x PDA medium (1 L)
19.5 g PDA
Pour the autoclaved medium after cooling it down to 50 °C - MS agar medium (1 L)
4.5 g MS and 7.5 g agar
Add 5 ml 50% (w/v) sucrose (filter sterilized) after cooling down the autoclaved medium to 50 °C
Mix well before pouring
Acknowledgments
Our research on fungal VCs has been supported by a grant to SK from the USDA-Specialty Crop Multi-State Program (AM170200XXXXG006), a fellowship to NL from the Storkan-Hanes-McCaslin Foundation, and the Huck Dissertation Research Award to NL. The I plate assay and the method for VC extraction were adopted from Ryu et al. (2003) and Stoppacher et al. (2010), respectively.
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Bailly, A. and Weisskopf, L. (2012). The modulating effect of bacterial volatiles on plant growth: current knowledge and future challenges. Plant Signal Behav 7(1): 79-85.
- Baldwin, I. T. (2010). Plant volatiles. Curr Biol 20(9): R392-397.
- Bennett, J. W., Hung, R., Lee, S. and Padhi, S. (2012). 18 Fungal and bacterial volatile organic compounds: An overview and their role as ecological signaling agents. In: Hock, B. (Ed.). Fungal Associations. Springer, Berlin, Heidelberg, 373-393.
- Bitas, V., Kim, H. S., Bennett, J. W. and Kang, S. (2013). Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol Plant Microbe Interact 26(8): 835-843.
- Bitas, V., McCartney, N., Li, N., Demers, J., Kim, J. E., Kim, H. S., Brown, K. M. and Kang, S. (2015). Fusarium oxysporum volatiles enhance plant growth via affecting auxin transport and signaling. Front Microbiol 6: 1248.
- Buck, L. B. (2004). Olfactory receptors and odor coding in mammals. Nutr Rev 62(11 Pt 2): S184-188; discussion S224-141.
- Herrmann, A. (2010). The chemistry and biology of volatiles. In: Herrmann, A. (Ed.). Chichester, John Wiley & Sons, Ltd.
- Hung, R., Lee, S. and Bennett, J. W. (2013). Arabidopsis thaliana as a model system for testing the effect of Trichoderma volatile organic compounds. Fungal Ecology 6(1): 19-26.
- Kai, M. and Piechulla, B. (2009). Plant growth promotion due to rhizobacterial volatiles--an effect of CO2? FEBS Lett 583(21): 3473-3477.
- Kováts, E. (1958). Gas-chromatographische charakterisierung organischer verbindungen. Teil 1: Retentionsindices aliphatischer halogenide, alkohole, aldehyde und ketone. Helvetica Chimica Acta 41(7): 1915-1932.
- Li, N., Alfiky, A., Wang, W., Islam, M., Nourollahi, K., Liu, X. and Kang, S. (2018a). Volatile compound-mediated recognition and inhibition between Trichoderma biocontrol agents and Fusarium oxysporum. Front Microbiol 9: 2614.
- Li, N. and Kang, S. (2018). Do volatile compounds produced by Fusarium oxysporum and Verticillium dahliae affect stress tolerance in plants? Mycology 9(3): 166-175.
- Li, N., Wang, W., Bitas, V., Subbarao, K., Liu, X. and Kang, S. (2018b). Volatile compounds emitted by diverse Verticillium species enhance plant growth by manipulating auxin signaling. Mol Plant Microbe Interact 31(10): 1021-1031.
- Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Pare, P. W. and Kloepper, J. W. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A 100(8): 4927-4932.
- Stoppacher, N., Kluger, B., Zeilinger, S., Krska, R. and Schuhmacher, R. (2010). Identification and profiling of volatile metabolites of the biocontrol fungus Trichoderma atroviride by HS-SPME-GC-MS. J Microbiol Methods 81(2): 187-193.
- Vaishnav, A., Kumari, S., Jain, S., Varma, A. and Choudhary, D. K. (2015). Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. J Appl Microbiol 119(2): 539-551.
- Xie, X., Zhang, H. and Pare, P. W. (2009). Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus subtilis (GB03). Plant Signal Behav 4(10): 948-953.
- Zhang, H., Kim, M. S., Krishnamachari, V., Payton, P., Sun, Y., Grimson, M., Farag, M. A., Ryu, C. M., Allen, R., Melo, I. S. and Pare, P. W. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226(4): 839-851.
- Zhang, H., Xie, X., Kim, M. S., Kornyeyev, D. A., Holaday, S. and Pare, P. W. (2008). Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J 56(2): 264-273.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, W., Li, N., Liu, X. and Kang, S. (2019). I Plate-based Assay for Studying How Fungal Volatile Compounds (VCs) Affect Plant Growth and Development and the Identification of VCs via SPME-GC-MS. Bio-protocol 9(4): e3166. DOI: 10.21769/BioProtoc.3166.
Category
Microbiology > Microbial metabolism > Other compound
Plant Science > Plant physiology > Plant growth
Biochemistry > Other compound > Small molecular
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










