- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Redox States of the β3 Integrin Disulfide Bonds by Mass Spectrometry
Published: Vol 9, Iss 3, Feb 5, 2019 DOI: 10.21769/BioProtoc.3156 Views: 5737
Reviewed by: Ralph Thomas BoettcherAruna BitraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Turnover Dynamics Analysis With Subcellular Spatial Resolution
Lorena Alamillo [...] Edward Lau
Aug 5, 2025 2397 Views

Immunopeptidomics Workflow for Isolation and LC-MS/MS Analysis of MHC Class I-Bound Peptides Under Hypoxic Conditions
Hala Estephan [...] Eleni Adamopoulou
Nov 20, 2025 1846 Views

Quantitative Proteomics of Nitrosylated Proteins in Melanoma Using the Biotin-Switch Technique Combined With Tandem Mass Tag Labeling
Vipin K. Yadav [...] Sanjay Premi
Dec 5, 2025 1539 Views
Abstract
Functional disulfide bonds mediate a change in protein function in which they reside when cleaved or formed. To elucidate how a functional disulfide bond controls protein activity, it is critical that the redox state of the bond in the population of protein molecules is known. Measurement of changes in disulfide bond redox state relies on thiol probes and immunoblotting. Such technique only offers a qualitative indication of a change in redox state but not the identity of cysteines involved. A differential cysteine alkylation and mass spectrometry technique is described here that affords precise quantification of protein disulfide bond redox state. The utility of the technique is demonstrated by quantifying the redox state of 24 of the 28 disulfide bonds in human β3 integrin from purified platelets.
Keywords: Disulfide bondsBackground
Disulfide bonds are covalent linkages between two cysteine residues in a protein. There are three types of disulfide bonds found in proteins: structural, catalytic and functional. Structural disulfide bonds stabilize protein conformation and are the most common type disulfide bonds. Catalytic disulfide bonds are found in redox enzymes such as thioredoxin and protein disulfide isomerases that reduce and oxidize disulfide bonds in substrate proteins. Functional disulfide bonds mediate a change of protein function such as activity, ligand affinity, localization or multimerization when cleaved or formed. Our understanding of how functional disulfides control protein function is limited by the molecular techniques available to detect changes in cysteines and disulfide bonds. Measurement of changes in disulfide redox state has mainly relied on the use of thiol probes such as biotin-conjugated maleimide (3-(N-maleimidylpropionyl)biocytin), followed by detection using streptavidin conjugated-peroxidase (Manickam et al., 2008). This provides a qualitative indication of a change in redox state and does not identify which cysteines are involved. The development of isotopic cysteine alkylators (unlabeled and 13Carbon labeled) has allowed differential cysteine alkylation, and mass spectrometry allows for an estimation of the fraction of a protein disulfide bond that is reduced in the protein preparation (Chambers et al., 2010).
In the method described herein, reduced disulfide bond cysteines are alkylated with 2-iodo-N-phenylacetamide (12C-IPA) and the oxidized disulfide bond cysteines with a stable carbon-13 isotope of IPA (13C-IPA) following reduction with dithiothreitol. The ratio of alkylation of peptides containing the disulfide bond cysteines with 12C-IPA compared to 13C-IPA represents the fraction of the disulfide in the population that is in the reduced state. The advantage of this pair of cysteine alkylators is that they have the same chemical reactivity and the same structure, which enhances the reliability of alkylation, resolution of the alkylated peptides by liquid chromatography and their detection by mass spectrometry (Pasquarello et al., 2004). A mass difference of 6 Da is the only change in a cysteine labeled with 12C-IPA or 13C-IPA.
To exemplify this technique, the redox state of disulfide bonds in β3 integrin from platelet is calculated. Based on the published crystal structure (PDB: 3FCS), there are 56 cysteines that form 28 disulfide bonds in β3 integrin. Using this protocol, sixty-eight IPA-labeled peptides encompassing 34 of the 56 cysteines in the β3 integrin subunit were resolved, which represents 24 of the 28 β3 integrin disulfide bonds. Among the 24 disulfide bonds measured, one disulfide bond, Cys177-184, is subjected to cleavage by disulfide-cleaving enzyme ERp5, thereby regulating ligand affinity of β3 integrin on platelet surface (Passam et al., 2018).
Materials and Reagents
- Ziptip® C18 pipette tip (Millipore, catalog number: ZTC18S96)
- 1.5 ml tubes
- Natural human αIIbβ3 protein purified from human platelets (Abcam, catalog number: ab95130)
- Anti-integrin β3 (GPIIIa, CD61) PSI domain [AP-3] antibody (Kerafast, catalog number: EBW106)
- 2-iodo-N-phenylacetamide (12C-IPA) (Cambridge isotopes, catalog number: ULM-8131-PK)
- 13C-2-iodo-N-phenylacetamide (13C-IPA) (Cambridge isotopes, catalog number: CLM-8130-0.1)
- Dimethyl sulfoxide (DMSO) (Sigma, catalog number: D8418)
- NuPAGETM 4x LDS sample loading buffer (Thermo Fisher Scientific, catalog number: NP0007)
- NuPAGETM 4-12% Bis-Tris gel 1.5 mm, 10-well (Thermo Fisher Scientific, catalog number: NP0335BOX)
- NuPAGETM MOPS running buffer 20x (Thermo Fisher Scientific, catalog number: NP0001)
- Brilliant Blue G solution (Sigma, catalog number: B8522-1EA)
- Ammonium bicarbonate (Sigma, catalog number: A6141)
- Dithiothreitol (DTT) (Sigma, catalog number: D0632)
- Formic acid LC-MS grade (Thermo Fisher Scientific, catalog number: 28905)
- Trifluoroacetic acid (Sigma, catalog number: 302031)
- Formic acid (Sigma, catalog number: F0507)
- Acetonitrile HPLC grade (Fisher Chemical, catalog number: 10407440)
- PNGase F Proteomics grade enzyme (Sigma, catalog number: P7367)
- Sequencing grade bovine chymotrypsin (Roche, catalog number: 11418467001)
- Trypsin Gold, Mass spectrometry grade (Promega, catalog number: V5280)
- HCl
- Acetic acid
- Calcium chloride
- Methanol
- Coomassie blue
- NaCl
- KCl
- Na2HPO4
- KH2PO4
- PBS (see Recipes)
- Gel destaining solution (see Recipes)
- 50 mM 12C-IPA/DMSO solution (see Recipes)
- 50 mM 13C-IPA/DMSO solution (see Recipes)
- 5 mM 13C-IPA/25 mM ammonium bicarbonate (see Recipes)
- 25 mM ammonium bicarbonate buffer (see Recipes)
- 25 mM ammonium bicarbondate/50% acetonitrile buffer (see Recipes)
- 25 mM ammonium bicarbonate/10 mM calcium chloride buffer (see Recipes)
- 1 M calcium chloride solution (see Recipes)
- 40 mM DTT/25 mM ammonium bicarbonate (see Recipes)
- 1 mM HCl solution (see Recipes)
- 50 mM acetic acid (see Recipes)
- 5% formic acid (see Recipes)
- 5% formic acid/50% acetonitrile buffer (see Recipes)
- 1% trifluoroacetic acid solution (see Recipes)
- Wetting solution (see Recipes)
- Equilibration solution (see Recipes)
- 0.1% formic acid (see Recipes)
- HPLC Buffer A (see Recipes)
- HPLC Buffer B (see Recipes)
Equipment
- Pipettes
- Scalpel
- Centrifuge
- Speedivac SC210P1-15 (Thermo Fisher Scientific)
- Thermomixer R with 1.5 ml block (Eppendorf, catalog number: Z605271)
- Q-Exactive Plus Dionex Ultimate 3000 (Thermo Fisher Scientific) or an equivalent Orbitrap mass spectrometer such as Velos (Thermo Fisher Scientific) or Q-Exactive Classic (Thermo Fisher Scientific)
- -20 °C freezer
- -80 °C freezer
- NanoDrop
Software
- RawConverter (free download at http://fields.scripps.edu/rawconv/) (He et al., 2015)
- Mascot search database (Matrix Science)
- Peptide/Protein MS Product Utility Program MS-Product from Protein Prospector Tools (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct)
- Qual Browser from XCalibur Software 2.2 (Thermo Fisher Scientific)
- Microsoft Excel (Microsoft) or equivalent spreadsheet tool
Procedure
- Differential cysteine labeling and in-gel digestion
Note: All washes that require mixing and incubations of samples are performed using Thermomixer R with a block that holds 1.5 ml tubes.- To 6 µg of natural human αIIbβ3, add PBS to 25 µl.
Note: For lyophilized proteins, a non-amide buffer with pH > 7.0 (e.g., PBS) is recommended for reconstitution. - Add 2.2 µl of 50 mM 12C-IPA/DMSO solution to final 4 mM. Mix and incubate at 25 °C for 1 h in the dark.
Note: Prepare fresh 12C-IPA/DMSO solution just before use. Discard any unused solution. Cysteine is a strong nucleophile, and its reaction with IPA should complete in 1 h. Extended incubation time with IPA is not recommended. - Add 9 µl of NuPAGETM 4x LDS sample loading buffer and heat samples at 70 °C for 10 min.
- Analyze samples on non-reducing NuPAGETM 4-12% Bis-Tris gel in 1x MOPS running buffer at 150 V for 65 min.
- Stain protein gel with 40 ml of Coomassie Blue G solution at room temperature for 30 min. Remove Coomassie Blue G solution.
- Destain gel in destaining solution at room temperature until background becomes clear.
- Excise protein gel bands corresponding to β3 integrin (Figure 1) with a clean scalpel.
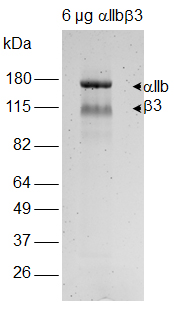
Figure 1. Analysis of αIIbβ3 integrin by non-reducing SDS-PAGE. Bands corresponding to β3 are excised for subsequent in-gel processing of samples. A photo of Coomassie blue stained gel is shown. - Dice gel band into small pieces of 1-2 mm3. Place gel pieces into a clean tube.
Note: Use standard 1.5 tubes that are nuclease-free. Special low protein binding tubes are not required. - Add 150 µl of 25 mM ammonium bicarbonate/50% acetonitrile buffer to gel pieces.
- Vortex gel pieces for 10 min at 25 °C using Thermomixer (Eppendorf). Remove supernatant by pipetting.
- Repeat Steps A9 and A10 for another 2 washes of the gel pieces.
- Dry gel pieces by adding 100 µl of 100% acetonitrile. Remove acetonitrile by pipetting.
Note: Gel pieces should be completely dried out within 1-2 min. Can store dried gel slices after this step at -20 °C. Long-term storage of dried gel slices is not recommended. However, the dried gel slices stored under this condition should remain stable for 1 month or longer. - Cover gel pieces with 80 µl of 40 mM DTT in 25 mM ammonium bicarbonate buffer. Incubate at 56 °C for 30 min. Remove supernatant.
- Wash three times with 150 µl of 25 mM ammonium bicarbonate/50% acetonitrile buffer. Vortex each time for 10 min at 25 °C.
- Dry gel slices by adding 100 µl of 100% acetonitrile. Remove acetonitrile by pipetting.
Note: It is not recommended to store gel pieces with proteins that contain free thiols generated from DTT reduction prior to alkylation with 13C-IPA. Gel pieces can be stored at -20 °C after alkylation with 13C-IPA, washed and dehydrated with 100% acetonitrile. - Prepare 50 mM 13C-IPA/DMSO solution and dilute it to 5 mM 13C-IPA in 25 mM ammonium bicarbonate buffer.
Note: Prepare fresh 13C-IPA/DMSO solution and dilute it to 5 mM 13C-IPA/25 mM ammonium bicarbonate solution just before use. 5 mM 13C-IPA/25 mM ammonium bicarbonate solution can precipitate slightly. Discard any unused solution. - Cover gel pieces with 100 µl of 5 mM 13C-IPA/25 mM ammonium bicarbonate solution and incubate in the dark for 1 h at 25 °C. Remove supernatant.
- Wash three times with 150 µl of 25 mM ammonium bicarbonate/50% acetonitrile buffer. Vortex each time for 10 min at 25 °C.
- Dry gel slices by adding 100 µl of 100% acetonitrile. Remove acetonitrile by pipetting.
- Dry gel pieces in Speedivac for 15-20 min to remove traces of acetonitrile prior to enzymatic processing of gel pieces.
- Reconstitute 50 units of PNGase F in 100 µl ddH2O.
Note: Any unused PNGase F can be stored at -20 °C. The enzyme remains stable for a few freeze-thawing cycles. For non-glycosylated proteins, skip PNGase F treatment (Steps A22-A28) and directly go to Step A29 to digest protein. - Add 10 µl (5 units) of the reconstituted PNGase F to dried gel pieces and incubate at 37 °C for 15-30 min.
Note: It is a very small volume to add to dried gel pieces, but it is not recommended to increase the volume of PNGase F by either diluting the enzyme or using more of the enzyme. Increased amount of PNGase F used can result in more PNGase F peptides that can distract the mass spectrometer in data acquisition. - Add 25 µl of water to the sample such that the gel pieces are just covered by the liquid.
- Incubate at 37 °C overnight.
- Centrifuge briefly to collect condensation. Remove and discard supernatant from the samples.
- Wash gel pieces with 250 µl of water and vortex at 25 °C for 20 min. Remove supernatant.
- Repeat Step A26 twice for 2 more washes.
- Dry gel pieces in Speedivac until gel pieces are completely dried.
Note: It may take up to 1 h to dry the gel pieces because of high water content. - Reconstitute chymotrypsin with 1 mM HCl to a final concentration of 1 mg/ml.
Note: Reconstituted chymotrypsin can be stored at 4 °C for 1 month. - Dilute chymotrypsin to 12.5 ng/µl in 25 mM ammonium bicarbonate/10 mM CaCl2 buffer.
- Add 35 µl of freshly diluted chymotrypsin to dried gel pieces and incubate at 37 °C for 4 h.
- Reconstitute trypsin with 50 mM acetic acid to a final concentration of 1 mg/ml.
Note: Reconstituted trypsin can be stored in 5 µl aliquots at -80 °C for over a year. - Dilute trypsin to 12.5 ng/µl in 25 mM ammonium bicarbonate buffer.
- Add 30 µl of freshly diluted trypsin to gel pieces and incubate at 25 °C overnight.
- Stop digestion of protein by adding 10 µl of 5% formic acid solution to sample.
- Transfer digest solution which contains peptides to a fresh tube.
- Elute remaining peptides from gel slices by adding 30 µl of 5% formic acid/50% acetonitrile solution to gel slices. Vortex for 20 min at 25 °C.
- Transfer supernatant to the tube containing peptides.
- Repeat Step A37 for another elution of peptides from gel pieces.
- Dry peptides with Speedivac with no heat applied.
- To 6 µg of natural human αIIbβ3, add PBS to 25 µl.
- Peptide clean up and mass spectrometry analysis
- Reconstitute peptides in 10 µl of 1% trifluoroacetic acid.
- Using a P10 pipet, wet the C18 Ziptip by aspirating 10 µl of wetting solution. Discard solution to waste.
Note: Avoid introducing air to the C18 matrix when pipetting. - Repeat Step B2 for a second time.
- Equilibrate Ziptip using 10 µl of Equilibration buffer. Discard solution to waste.
- Repeat Step B4 for another two times.
- Bind peptides by aspirating 10 µl of the reconstituted peptide sample. Aspirate up and down no more than 4 times.
Note: Do not aspirate more than 4 times as this will select for high-affinity peptides and lose peptides that only weakly bind to C18 matrix. - Wash column by aspirating 10 µl of Equilibration buffer. Discard wash to waste.
- Repeat Step B7 for another 2 times.
- Elute bound peptides using 5 µl of wetting solution by aspirating up and down 3 times in a clean tube.
- Dry peptides in Speedivac with no heat applied.
- Reconstitute peptides in 12 µl of 0.1% formic acid.
- Measure the concentration of peptide using a NanoDrop. Use 0.1% formic acid as blank.
- Dilute the peptide to 80 ng/µl with 0.1% formic acid.
- Inject 3 µl of peptides (200-250 ng) into a Thermo Scientific Dionex Ultimate 3000 and analyze peptides on a 35 cm x 75 μm C18 reverse phase analytical column with integrated emitter using a gradient of 2%-35% acetonitrile over 22 min (Table 1) with a flow rate of 300 nl/min.
Note: Mass spectrometry analysis is best to be performed at a proteomic facility where technical support can be provided to set up the method to run peptide samples. The intensity of the TIC peaks should be in the range of 1E9-1E10.
Table 1. Solvent gradient used in LC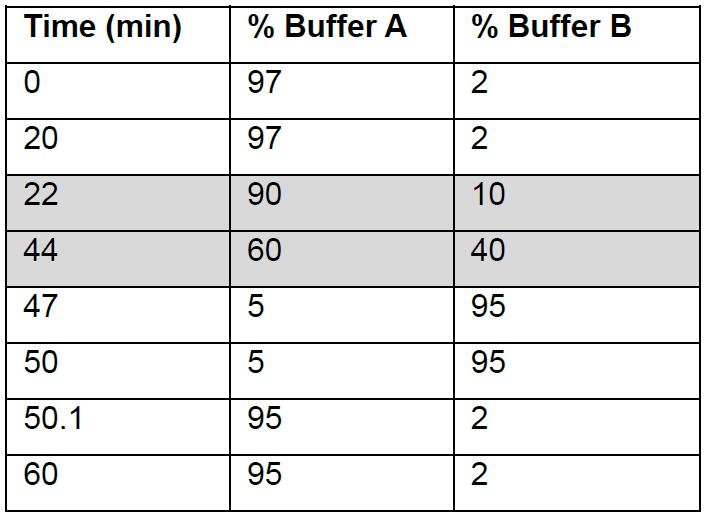
Acetonitrile gradient for peptide elution is highlighted. - Tandem mass spectrometry (MS/MS) analysis is performed on a Q-Exactive Plus mass spectrometer (Thermo Scientific) using HCD fragmentation. Set electrospray ionization of peptides at +2.0 kV. The scan range for full MS is set at 350 to 1,750 m/z. The spectrum data type for full MS is profile mode whereas for MS/MS is centroid mode.
- Data-dependent acquisition method is used to obtain MS/MS spectra of the top 10 most abundant ions at any one point during the gradient.
Data analysis
- Identification of peptides using Mascot search engine
- Data files generated from LC-MS/MS (.raw) are converted to mascot generic format (.mgf) using RawConverter (He et al., 2015).
- Using Mascot Daemon, set parameters under Parameter Editor.
- Taxonomy: All entries.
- Database: Swissprot.
- Variable modification: click select modification and show all modifications. Select iodoacetanilide derivative (C), iodoacetanilide 13C derivative (C), oxidation (M) and deamidation (NQ).
- Variable modifications, select all modifications, then choose iodoacetanilide derivative (C), iodoacetanilide 13C derivative (C), oxidation (M) and deamidation (NQ).
- Select enzymes as TrypChymo.
- Select Max. Missed cleavages to 3.
- Select monoisotopic for mass.
- Select peptide charge 2+, 3+ and 4+.
- Set peptide tolerance at ± 6 ppm and MS/MS tolerance at ± 0.6 Da.
- Select MS/MS ions search.
- Data format as Mascot generic.
- Save parameter settings as beta3_integrin.par.
- Under Task Editor, add .mgf files to be analyzed by mascot search and set parameter by choosing beta3_integrin.par file. Then start the search by pressing the Run button.
- Once the search is completed, click the hyperlink for Results file URL to bring up the search data. To obtain sufficient cysteine-containing peptides for quantification of the redox state of disulfide bonds, it will require 65%-80% coverage of β3 integrin.
- Identify all cysteine-containing peptides from mascot search and tabulate them in a spreadsheet in Microsoft Excel.
- Calculating m/z ratios of cysteine-containing peptides
- Using the list of cysteine-containing peptides identified by Mascot, calculate their monoisotopic mass/charge ratio (m/z) of +2, +3 and +4 using Peptide/Protein MS Product Utility Program MS-Product.
- Cys-iodoacetanilide has a mass adduct of 133.05276, and Cys-13C-iodoacetanilide has a mass adduct of 139.07289, which are differed by 6 Da due to the six 13C-carbon ring on IPA (Table 2).
- To calculate the m/z ratios of the peptide, for example, ISPPEALENPCY from β3, enter ISPPEALENPC(133.05276)Y and ISPPEALENPC(139.07289)Y as input sequences in MS-Product from Protein Prospector for 12C-IPA and 13-IPA labeled peptides, respectively.
- Set charge to 4 to obtain values of m/z ratios from +1 to +4. Values for m/z ratios should be at least 4 decimal places (Table 3).
- Tabulate the m/z ratios (+2, +3 and +4) in the same spreadsheet that recorded all cysteine-containing peptides identified.
Table 2. Monoisotopic mass of adduct in cysteine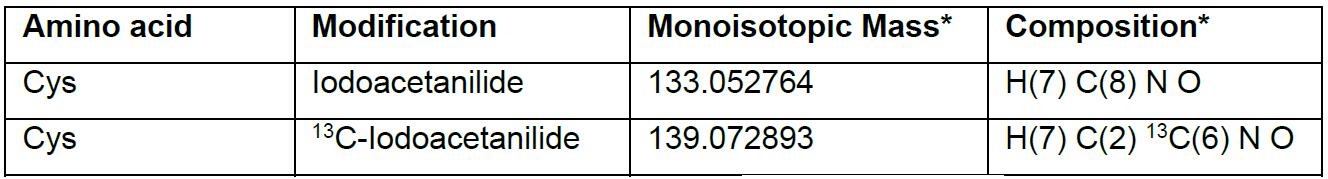
*Values and composition are obtained from Unimod (www.unimod.org/).
Table 3. Monoisotopic m/z ratios for ISPPEALENPCY peptide from β3 integrin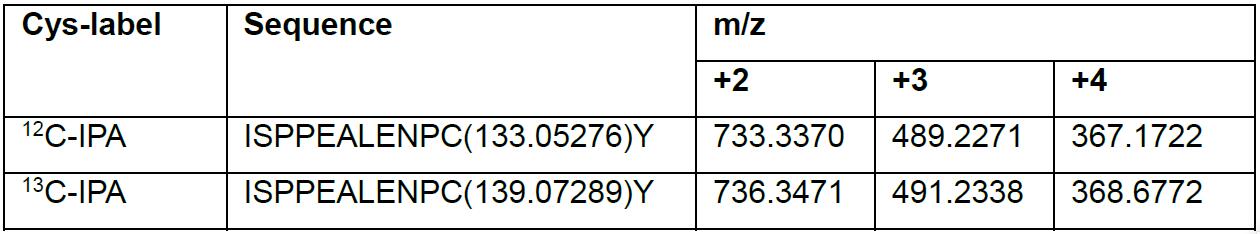
- Quantification of the abundance of single cysteine-containing peptide using XCalibur Qual Browser
- To calculate ion abundance of peptides, extracted ion chromatograms were generated using the XCalibur Qual Browser software. Set autoprocessing parameters and display options for chromatogram and spectra as follows:
- Activate the chromatogram cell (top panel) by selecting the pin
 at the upper right corner. To define parameters for autoprocessing for chromatograms, choose Display and then Ranges from the drop down menu. When the Ranges dialog box opens, select Autoprocessing and click Use user defined for Mass Tolerance and set it as 6 ppm. Mass precision is set at 5 Decimals. For display options, choose Display and then Display Options. Select Labels and choose Retention time (2 Decimals) and Area.
at the upper right corner. To define parameters for autoprocessing for chromatograms, choose Display and then Ranges from the drop down menu. When the Ranges dialog box opens, select Autoprocessing and click Use user defined for Mass Tolerance and set it as 6 ppm. Mass precision is set at 5 Decimals. For display options, choose Display and then Display Options. Select Labels and choose Retention time (2 Decimals) and Area. - Activate the spectrum cell (bottom panel) by selecting the pin
 at the upper right corner. To define parameters for autoprocessing for spectra, choose Display and then Ranges from the drop down menu. When the Ranges dialog box opens, select Autoprocessing and click Use user defined for Mass Tolerance and set it as 6 ppm. Mass precision is set at 5 Decimals. For display options, choose Display and then Display Options. Select Labels and choose Mass (5 Decimals).
at the upper right corner. To define parameters for autoprocessing for spectra, choose Display and then Ranges from the drop down menu. When the Ranges dialog box opens, select Autoprocessing and click Use user defined for Mass Tolerance and set it as 6 ppm. Mass precision is set at 5 Decimals. For display options, choose Display and then Display Options. Select Labels and choose Mass (5 Decimals). - To save the settings, select File and then Layout from the drop down menu, choose Save as Default. The above settings for autoprocessing and display will be used for subsequent quantification of MS peaks.
- Activate the chromatogram cell (top panel) by selecting the pin
- Use the m/z ratios (+2, +3, +4) calculated from MS-Product to search ions containing the corresponding m/z values.
- Select the chromatogram cell and choose Display then Ranges from the drop down menu.
- When the Ranges dialog box opens, in the Plot properties, choose Scan filter FTMS + p NSI Full mass [350.00-1750.00] from the drop down menu.
- Enter the values for +2, +3 and +4 m/z in Ranges.
- Click the automated peak detection function
 built into the program to calculate peak area (Figure 2).
built into the program to calculate peak area (Figure 2). - Record integrated peak area in the Microsoft Excel spreadsheet once verified (see Step C3).
- Automated peaks from Qual Browser are verified by the followings.
- The retention time of Cys-iodoacetanilide or Cys-13C-iodoacetanilide labeling of the same peptide should be identical since 12C-IPA and 13C-IPA are identical in their chemical properties, and hence the two species of peptides of the same sequence should resolve on the column in identical fashion (Figure 2).
- The retention time of a peptide identified by Mascot database search and Qual Browser should be of similar value and should not be differed by more than 0.02-0.03 min.
- Compare the retention time from Mascot search which is in seconds (rtinseconds) to verify peaks in the Qual Browser.
- Check the isotopic distribution of the m/z ratios (+2 or +3 or both). For m/z ratio of +2, the isotopic peaks should differ by 0.5, and for m/z ratio of +3, the isotopic peaks should differ by 0.33 (Figure 2).
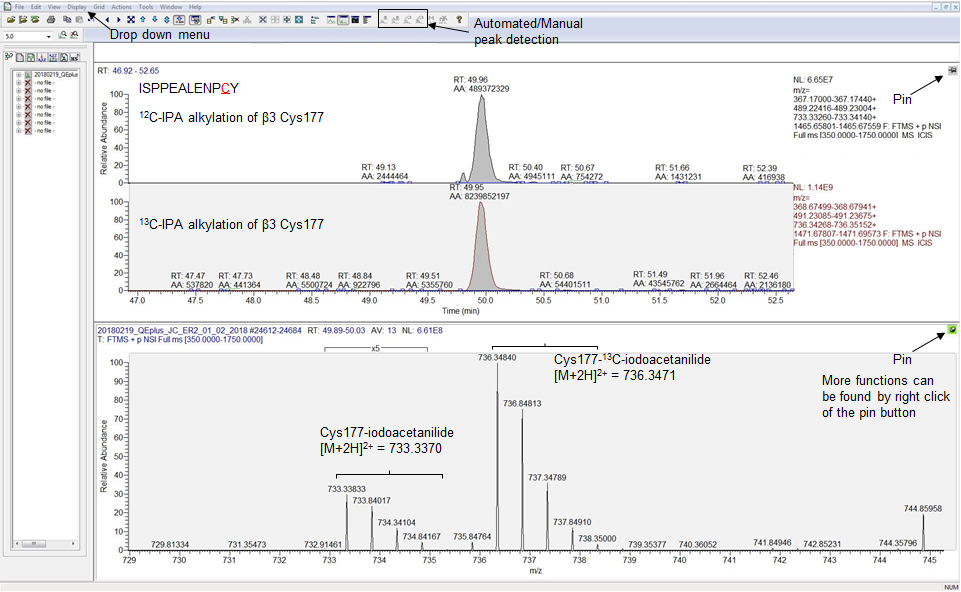
Figure 2. Screenshot of the XCalibur Qual Browser Window showing automated peaks detected for the peptide ISPEALEPCY labeled with 12C-IPA or 13C-IPA. The two peptide species have identical retention times, and the automated area is used for quantification of their abundance. The automated peaks correspond to m/z of 733.33833 and 736.34840 for ISPPEALENPC(133.05276)Y and ISPPEALENPC(139.07289)Y respectively, with an isotopic mass distribution of 0.5 confirming their m/z ratios as +2. Utilities such as Drop down menu, pin button, and automated peak detection are shown by black arrows.
- The fraction of reduced disulfide bond is expressed by the area for Cys-iodoacetanilide labeled peptide over the sum of areas for Cys-iodoacetanilide and Cys-13C-iodoacetanilide labeled peptides.
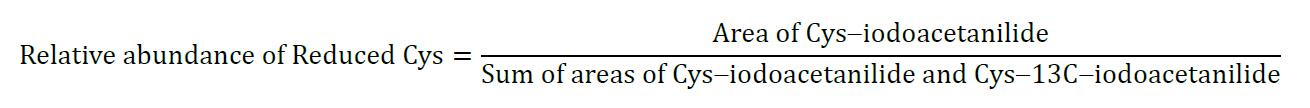
- To calculate ion abundance of peptides, extracted ion chromatograms were generated using the XCalibur Qual Browser software. Set autoprocessing parameters and display options for chromatogram and spectra as follows:
- Quantification of the abundance of two cysteine-containing peptide using XCalibur Qual Browser
- For peptides containing two cysteines such as YCNCTTR from β3, alkylation with 12C-IPA and 13C-IPA will give rise to four species of the same peptide; fully reduced, partially reduced and fully oxidized (Table 4).
- There are two species for the partially reduced peptide since it can be a combination of reduced Cys1 and oxidized Cys2, or vice versa. Both species of partially reduced peptide have identical m/z ratios. Hence the area generated by Qual browser for partially reduced peptide is the sum of both species.
- To delineate their percentage of abundance, b and y ions derived from peptide fragmentation that are unique to Cys1 and Cys2 are searched from the tandem mass spectrum, and their intensities are used for quantification.
- The mass for b and y ions of YC(133.05276)NC(139.07289)TTR and YC(133.09.07289)NC(133.05276)TTR can be calculated using protein prospector (Table 5).
- Tabulate the mass for unique b and y ions using a Microsoft Excel spreadsheet.
Table 4. Monoisotopic m/z ratios for the β3 integrin YCNCTTR peptide alkylated with 12C-IPA and 13C-IPA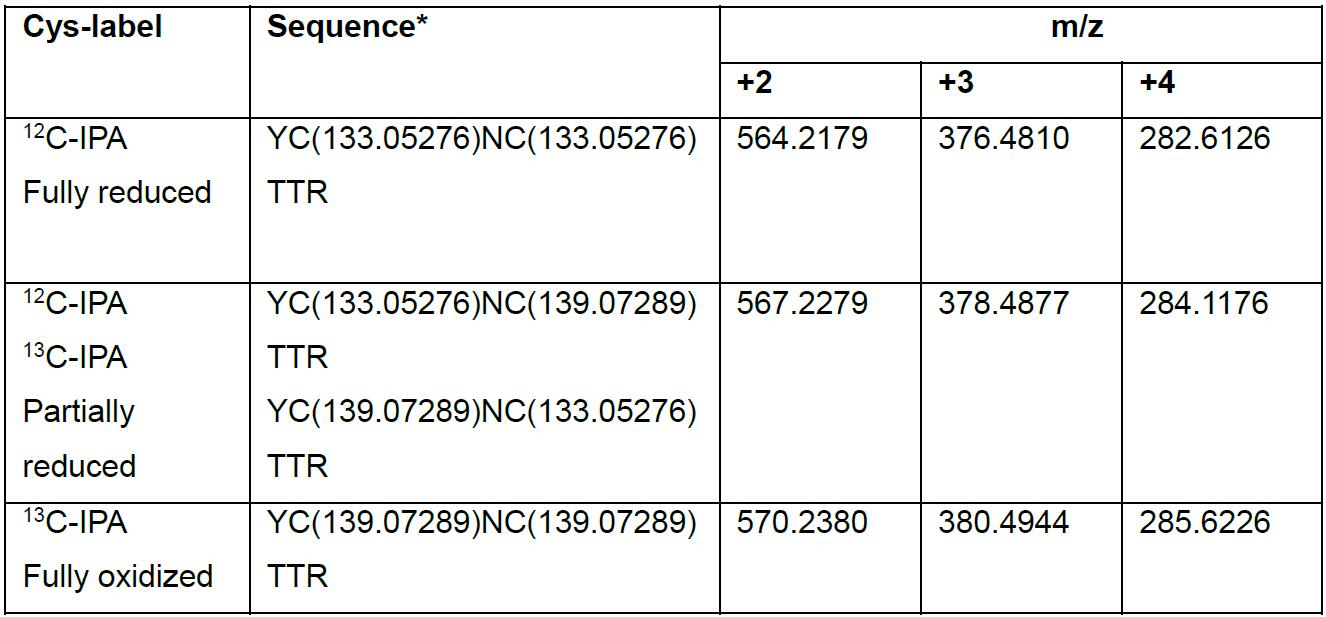
*YCNCTTR is a glycosylated peptide where N becomes deamidated after PNGase F treatment, and the actual peptide should be YCN(deamidated)CTTR.
Table 5. Values of b and y ions generated from partially reduced peptide YCNCTTR*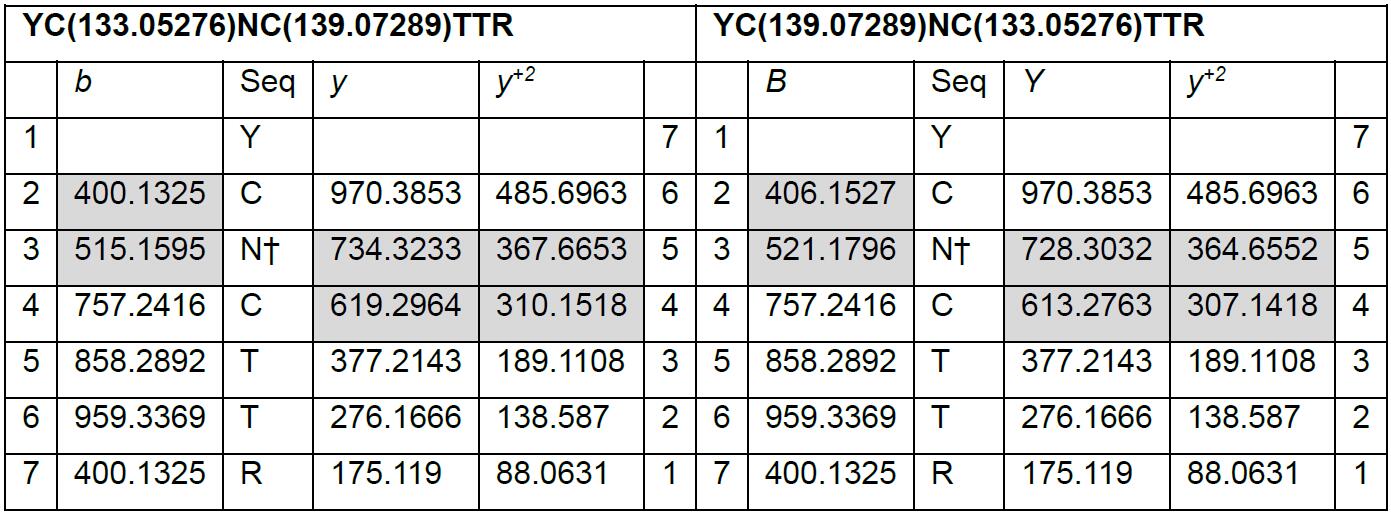
*Unique b and y ions are highlighted in grey. Values for b and y ions are obtained from MS Product tool in Protein Prospector.
†N is a glycosylation site and becomes deamidated post PNGase F treatment. - The MS/MS spectrum for the partially reduced YCNCTTR peptide is then used for quantification.
- To bring up the tandem mass spectrum for the partially reduced YCNCTTR peptide, select chromatogram cell to make it active and bring up the Ranges dialog box.
- In the Plot properties, choose TIC as plot type and in the scan filter drop down menu, select FTMS + c NSI d Full ms2 567.22, which is the m/z ratio of +2. Some peptides may require m/z ratio of +3 to search for the tandem spectrum. If the exact m/z ratio is not available from the menu, select one that is within the range of ±0.5.
- On the chromatogram cell, select the ion peak that is within 1 min of the retention time of the automated peak area generated from the full scan (Figure 3).
- Select spectrum cell to make it active and the tandem mass spectrum corresponding to the ion peak selected on the chromatogram cell will be displayed. If the scan # of the tandem spectrum is known (from Mascot search), apply Default Layout and search for ion peak with the scan # from the chromatogram cell to display the tandem spectrum in the spectrum cell.

Figure 3. Screenshot of the XCalibur Qual Browser Window for quantification of YCNCTTR peptide labeled with Cys-iodoacetanilide and Cys-13C-iodoacetanilide. Automated peak detected for m/z = 567.2279 (mass range 567.22450-567.23130), ion peak (TIC) with m/z = 567.2192 and the corresponding tandem mass spectrum (scan # 20763) are shown. Unique b and y ions for Cys1 and Cys2 are found in the spectrum (red labels and arrows). The intensities of the fragment ions can be exported for quantification.
- To view and export the intensity values of all ion masses.
- Select View from the drop down menu, then choose Spectrum list. The ions masses and their exact and relative intensities will be shown.
- To export the intensity data, right click the mouse on the pin button, select Export then Clipboard (Exact mass).
- Paste data to a spreadsheet in Microsoft Excel.
- To calculate percentage of each species of the partially reduced peptide:
- Using the filter function in Microsoft Excel, search for masses of all unique b and y ions (values rounded to whole digit) and record their intensities.
- The relative abundance of the two partially reduced peptide species is expressed as:

- The total abundance of the reduced form for each cysteine of the peptide is the sum of the areas for fully reduced cysteine and that of partially reduced form of cysteine. Its relative abundance is expressed as a percentage of the Cys-iodoacetanilide to the sum of Cys-iodoacetanilide and Cys-13C-Iodoacetanilide.
- Cysteine containing peptides identified by differential cysteine alkylation and mass spectrometry can be found in (Passam et al., 2018, supplementary file 1). A total of 68 cysteine-containing peptides were identified by this methodology with 35 cysteines representing 24 of the 28 disulfide bonds in β3 integrin (Passam et al., 2018) which is 87% coverage of β3 disulfide bonds.
Note: Unmapped cysteines are found in peptides containing 3-5 cysteines and are beyond the limit of the current approach. Other proteomic approaches are necessary to overcome this limitation.
- For peptides containing two cysteines such as YCNCTTR from β3, alkylation with 12C-IPA and 13C-IPA will give rise to four species of the same peptide; fully reduced, partially reduced and fully oxidized (Table 4).
Notes
- Redox state of disulfides in αIIbβ3 will slightly vary between batches of purified αIIbβ3, but differences are generally non-significant.
- To measure the redox state of disulfide bonds in β3 integrin on platelet surface, free cysteines on the surface of wash platelets are labeled with 5 mM 12C-IPA in PBS for 30 min in the dark. β3 integrin is then subjected to immunoprecipitation as described in Passam et al. 2018. Eluates were analyzed by SDS-PAGE, followed by excision of gel band, DTT reduction, and 13C-IPA labeling as described in this protocol. 12C-IPA is also cell-permeable and therefore can be used to label free cysteine thiols in proteins that reside in a cell. Cells are lysed in buffered containing 5 mM 12C-IPA, and the protein of interested is purified by immunoprecipitation.
Recipes
- PBS
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g of KH2PO4
Adjust the pH to 7.4 with HCl
ad 1,000 ml ddH2O - Gel destaining solution
100 ml methanol (10%)
50 ml acetic acid (5%)
ad 1,000 ml ddH2O - 50 mM 12C-IPA/DMSO solution (freshly prepared)
13.055 mg 12C-IPA
ad 1 ml DMSO - 50 mM 13C-IPA/DMSO solution (freshly prepared)
13.055 mg 13C-IPA
ad 1 ml DMSO - 5 mM 13C-IPA/25 mM ammonium bicarbonate
1 ml 50 mM 13C-IPA/DMSO solution
ad 10 ml 25 mM ammonium bicarbonate buffer - 25 mM ammonium bicarbonate buffer
1.9764 g ammonium bicarbonate
ad 1,000 ml ddH2O - 25 mM ammonium bicarbondate/50% acetonitrile buffer
1.9764 g ammonium bicarbonate
500 ml acetonitrile
ad 1,000 ml ddH2O - 25 mM ammonium bicarbonate/10 mM calcium chloride buffer (freshly prepared)
990 µl 25 mM ammonium bicarbonate buffer
10 µl 1 M calcium chloride solution
ad 1 ml ddH2O - 1 M calcium chloride solution
5.549 g calcium chloride
ad 50 ml ddH2O - 40 mM DTT/25 mM ammonium bicarbonate (freshly prepared)
61.7 mg DTT
ad 10 ml ddH2O - 1 mM HCl solution
100 µl 1 M HCl
ad 100 ml ddH2O - 50 mM acetic acid
285.7 µl glacial acetic acid (99.9%)
ad 100 ml ddH2O - 5% formic acid
5 ml formic acid
ad 100 ml ddH2O - 5% formic acid/50% acetonitrile buffer
5 ml formic acid (> 95%)
50 ml acetonitrile
ad 100 ml ddH2O - 1% trifluoroacetic acid solution
1 ml trifluoroacetic acid (99.9%)
ad 100 ml ddH2O - Wetting solution (0.1% trifluoroacetic acid/50% acetonitrile)
10 ml 1% trifluoroacetic acid
50 ml acetonitrile
ad 100 ml ddH2O - Equilibration solution (0.1% trifluoroacetic acid/5% acetonitrile)
10 ml 1% trifluoroacetic acid
5 ml acetonitrile
ad 100 ml ddH2O - 0.1% formic acid
100 µl formic acid (> 95%)
ad 100 ml ddH2O - HPLC Buffer A
95% LC-grade water
5% acetonitrile
0.1% formic acid
ad 1,000 ml ddH2O - HPLC Buffer B
20% LC-grade water
80% acetonitrile
0.1% formic acid
ad 1,000 ml ddH2O
Acknowledgments
This work was supported by the Helen and Robert Ellis Fellowship and Tony Basten Postdoctoral Fellowship from the Sydney Medical School Foundation. This work is adapted from previous work (Passam et al., 2018).
Competing interests
The author declares no conflict of interest.
References
- Chambers, J. E., Tavender, T. J., Oka, O. B. V. Warwood, S., Knight, D. and Bulleid, N. J. (2010). The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1α. J Biol Chem. 285 (38): 29200-29207.
- He, L., Diedrich, J., Chu, Y. Y. and Yates, J. R., 3rd (2015). Extracting accurate precursor information for tandem mass spectra by rawconverter. Anal Chem 87(22): 11361-11367.
- Manickam, N., Sun, X., Hakala, K. W., Weintraub, S. T. and Essex, D. W. (2008). Thiols in the αIIbβ3 integrin are necessary for platelet aggregation. Br J Haematol 142(3): 457-465.
- Pasquarello, C., Sanchez, J. C., Hochstrasser, D. F. and Corthals, G. L. (2004). N-t-butyliodoacetamide and iodoacetanilide: two new cysteine alkylating reagents for relative quantitation of proteins. Rapid Commun Mass Spectrom 18(1): 117-127.
- Passam, F., Chiu, J., Ju, L., Pijning, A., Jahan, Z., Mor-Cohen, R., Yeheskel, A., Kolsek, K., Tharichen, L., Aponte-Santamaria, C., Grater, F. and Hogg, P. J. (2018). Mechano-redox control of integrin de-adhesion. Elife 7: e34843.
Article Information
Copyright
Chiu. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chiu, J. (2019). Measurement of Redox States of the β3 Integrin Disulfide Bonds by Mass Spectrometry. Bio-protocol 9(3): e3156. DOI: 10.21769/BioProtoc.3156.
- Passam, F., Chiu, J., Ju, L., Pijning, A., Jahan, Z., Mor-Cohen, R., Yeheskel, A., Kolsek, K., Tharichen, L., Aponte-Santamaria, C., Grater, F. and Hogg, P. J. (2018). Mechano-redox control of integrin de-adhesion. Elife 7: e34843.
Category
Biochemistry > Protein > Quantification
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










