- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Induction and Analysis of Anti-CD40-induced Colitis in Mice
Published: Vol 9, Iss 3, Feb 5, 2019 DOI: 10.21769/BioProtoc.3153 Views: 11058
Reviewed by: Ruth A. FranklinWathsala WijayalathAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2360 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 1984 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2232 Views
Abstract
Colon inflammation or colitis affects more than 1 million people worldwide. Several pre-clinical models, including chemical-induced (i.e., DSS, TNBS) or pathogen-induced (i.e., Citrobacter rodentium) have been used to study mechanisms involved in the development and regulation of colitis. Anti-CD40 induced colitis model has gained acceptance to study the roles of innate immune cells during acute intestinal inflammation. Here we describe a rapid, robust and reproducible protocol to induce and analyze anti-CD40 mediated colitis in mice.
Keywords: Anti-CD40Background
Inflammatory Bowel Disease (IBD), including Crohn’s disease and Ulcerative colitis, affects about 1.5 million people in the United States (Ng et al., 2017). To better understand the mechanisms involved in the development and progression of IBD, a number of pre-clinical models (i.e., DSS, TNBS, anti-CD40, etc.) have been developed to address various aspects of immune response during tissue injury over the last two decades. CD40 is highly expressed by the colon lamina propria antigen presenting cells. We have demonstrated that activation of CD40 signaling using an agonist anti-CD40 antibody can trigger colitis in T and B cells deficient mice (here referred as Rag-/- mice) driven by excessive production of IL-23, IL-1β and IL-12 by myeloid cells (Uhlig et al., 2006). Anti-CD40 model is a unique model of colitis driven by IL-23-producing gut resident CX3CR1+ macrophages and IL-22-producing group 3 innate lymphoid cells (ILC3) (Bauche et al., 2018). This model of colitis is restricted to the proximal colon and is a potent model to study the role of innate immunity in colon inflammation. Here, we describe a robust and reproducible method to induce and analyze anti-CD40-induced colitis in mice. Anti-CD40-treated Rag2-/- mice lose up to 20% of their initial weight within three days post injection, and then return to their initial weight by Day 7 post induction (Figure 1A). Elevated levels of pro-inflammatory cytokines can be detected in the proximal colon as soon as Day 1 post induction (Cayatte et al., 2012) (Figure 1B) but maximal disease–characterized by massive infiltration of innate immune cells, loss of goblet cells and development of mitotic figures–is observed in the proximal colon at day 7 post disease induction (Figure 1C). Immune cell infiltration in the proximal colon and cytokine production, such as IL-22, by innate lymphoid cells can be measured by flow cytometry (Figure 1D).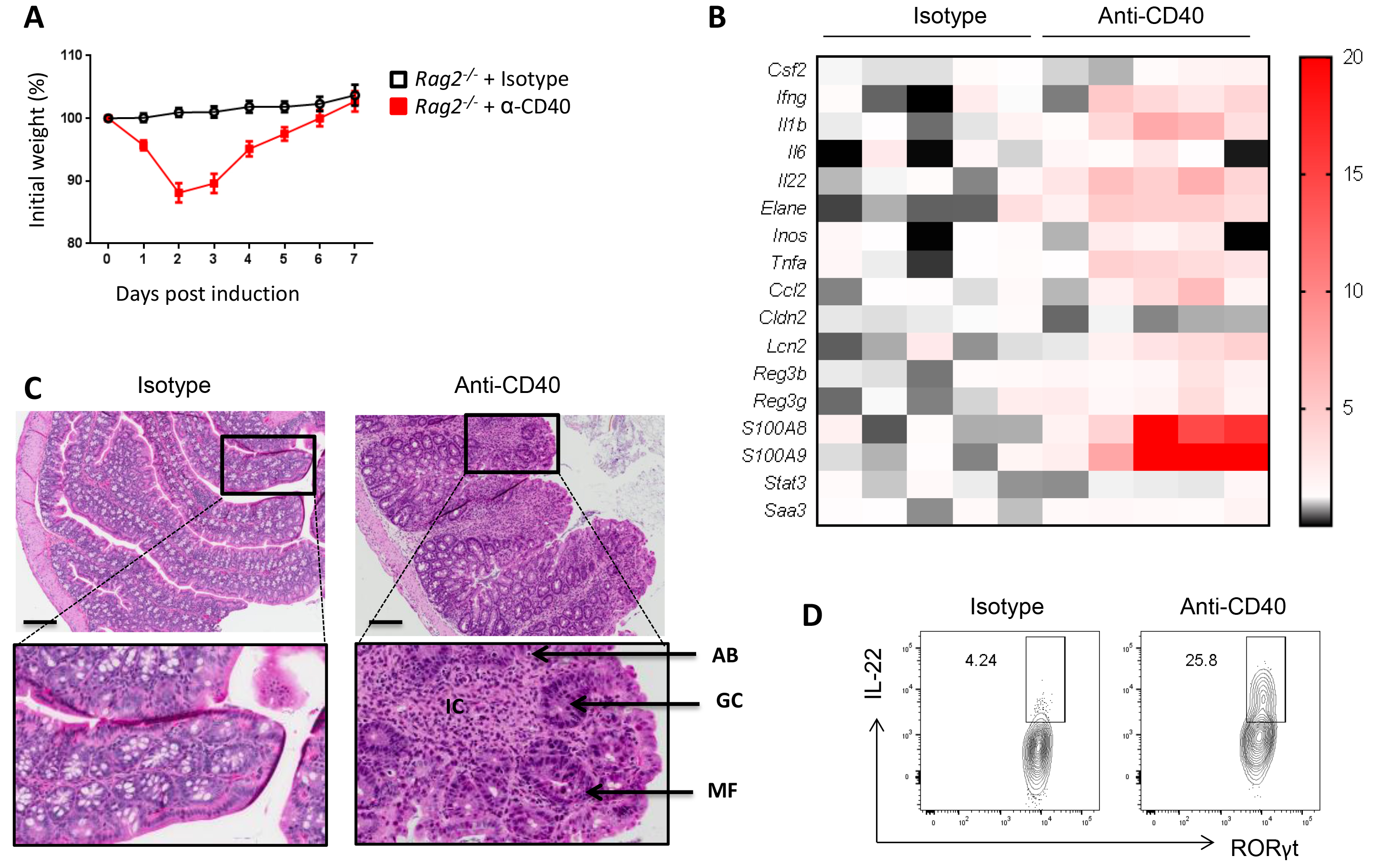
Figure 1. Induction and analysis of the anti-CD40 colitis mouse model. A. Percentage of initial weight over a 7 days period. Initial weight is measured right before injection of isotype or anti-CD40 antibodies. B. Gene expression profile of the proximal colon at Day 7 post treatment. Data shows relative fold change over the isotype control. C. Representative photomicrographs of H&E stained colon section 7 days after injection of isotype (left) or anti-CD40 antibodies (right). IC: Immune cell infiltration; GC: Loss of Goblet cells; AB: Apoptotic body; MF: Mitotic figures. Scale bars = 250 μm. D. Representative dot plot of IL-22 production by proximal colon lamina propria ILC3 (gated on lineage-, CD90high, CD45int, RORγt+ cells) at Day 2 post treatment.
Materials and Reagents
- Materials
- Pipette tips (Thermo Scientific)
- Eppendorf tubes, 1.5 ml (Eppendorf, catalog number: 05-402-5)
- 1 ml sterile sub-Q syringe 26 G (BD, catalog number: 3095971)
- Sterile cell strainer 70 μm Nylon mesh (Fisher Scientific, catalog number: 22363548)
- 50 ml centrifuge tube (Fisher Scientific, catalog number: 05-539-13)
- 15 ml conical centrifuge tube (Fisher Scientific, catalog number: 339650)
- 15 ml sterile plastic tube (Fisher Scientific, catalog number: 14-956-1D)
- 5 ml Serological pipettes (Falcon, catalog number: 357543)
- 25 ml Serological pipettes (Fisher Brand, catalog number: 13-678-11)
- Petri dishes (Falcon, catalog number: 351029)
- Serum or plasma separation tubes (SARSTEDT, catalog number: 41.1378.005)
- Parafilm (Pechiney, catalog number: PM-996)
- V bottom 96-well plate (Costar, catalog number: 3894)
- 70 μm cell strainer
- Animals
Eight to twelve weeks old Rag2-/- mice (Taconic, catalog number: RAGN12) were used to study anti-CD40 mediated colitis. - Reagents
- Liquid nitrogen
- Anti-mouse CD45 APC-eFluor780 antibody (eBioscience, catalog number: 47-0451-82)
- Anti-mouse CD90.2 BV786 antibody (BD Bioscience, catalog number: 564365)
- Anti-mouse RORγt PerCP-eFluor710 antibody (eBioscience, catalog number: 46-6981-82)
- Anti-mouse NK1.1 Alexa Fluor 700 antibody (BD Bioscience, catalog number: 560515)
- Anti-mouse CD11c PE-Cy7 antibody (BD Bioscience, catalog number: 561022)
- Anti-mouse CD11b FITC antibody (BD Bioscience, catalog number: 557396)
- Anti-mouse IL-22 APC antibody (eBioscience, catalog number: 17-7222-82)
- Endotoxin-free Anti-CD40 antibody (Clone FKG45, BioXCell, catalog number: BE0016-2)
- Anti-Rat IgG2a isotype control (Clone 2A3, BioXCell, catalog number: BP0089)
- HyClone Phosphate Buffered Saline (DPBS), 1x (GE Healthcare, catalog number: SH30028.02)
- Hank’s buffered salt solution (HBSS)
- 10% Neutral Buffered Formalin (Thermo Fisher Scientific, catalog number: 245-685)
- Penicillin/streptomycin solution 100x (Corning, catalog number: 30-002C1)
- Sodium pyruvate 100 mM (Gibco, catalog number: 11360070)
- 2-Mercaptoethanol 55 mM (Gibco, catalog number: 21985023)
- Ultra Pure 0.5 μM EDTA pH 8.0 (Gibco, catalog number: 15575-038)
- HEPES buffer solution 1 M (Gibco, catalog number: 15630-080)
- Hyclone Standard Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, catalog number: SH30088.03)
- Collagenase type VIII (Sigma-Aldrich, catalog number: C2139)
- DNase I (Sigma-Aldrich, catalog number: D5025)
- Dispase (Corning, catalog number: 354235)
- Percoll (GE Healthcare, catalog number: 17-0891-01)
- IMDM, GlutaMAX supplement (Thermo Fisher Scientific, catalog number: 31980097)
- Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, catalog number: P8139)
- Ionomycin (Sigma-Aldrich, catalog number: I0634)
- Brefeldin A (BD Bioscience, catalog number: 555029)
- Stain Buffer (BD, Pharmingen, catalog number: 554656)
- Transcription Factor Buffer Set (BD, Pharmingen, catalog number: 562725)
- Fixable viability stain 510 (BD, Horizon, catalog number: 564406)
- Mouse BD Fc Block (BD, Pharmingen, catalog number 553141)
- Ethanol 70% (Fisher Scientific, catalog number: HC1500)
- Ethanol 95% (Fisher Scientific, catalog number: HC1300)
- Ethanol 100% (Fisher Scientific, catalog number: HC600)
- Xylene (Fisher Scientific, catalog number: HC700)
- Paraffin (Fisher Scientific, catalog number: 23-0210-400)
- Hematoxylin (MasterTech Scientific, catalog number: HXHHEGAL)
- Eosin (Thermo Scientific, catalog number: 71311)
- Collagenase D (Sigma, catalog number: C5138-1G)
- Epithelial cell dissociation solution (see Recipes)
- Enzyme digestion mix (see Recipes)
- Complete IMDM media (see Recipes)
Equipment
- Scalpel (Southmedic, catalog number: SMI1/73-0121)
- Pipettes (Thermo Scientific)
- Forceps and Scissors (Fine Science Tools)
- Sterile cell culture hood
- Centrifuge (Thermo Fisher Scientific)
- Shaking water bath (Precision)
- Brightfield Microscope
- LSRII Flow cytometer (BD Biosciences)
- Incubator
- Vortex (Scientific Industries)
Software
- FlowJo_V10 (FlowJo, LLC, https://www.flowjo.com)
- GraphPad Prism 7 (GraphPad, https://www.graphpad.com)
- Microsoft Excel (Microsoft)
Procedure
- Induction of colitis in Rag2-/- mice
- Dilute the anti-CD40 antibody or isotype control to 1 mg/ml in sterile 1x PBS.
- Inject 100 μg of anti-CD40 or isotype control antibody intraperitoneal (i.p.) or 50 μg intravenous injection (i.v.)–based on researcher’s technical expertise–with a 1 ml sterile sub-Q syringe 26 G. Each batch of anti-CD40 antibody should be verified to be endotoxin-free (by checking with the manufacturer) to avoid systemic immune activation.
Notes:- Every animal facility will have a specific microbial environment, so it is crucial to determine the dose of anti-CD40 empirically by performing an initial dose-titration (25-100 μg) (Figure 2).
- A dose that is too high will result in death of the animals while an insufficient dose will lead to weak disease induction (characterized by inconsistent or poor colon inflammation) and variability in colitis development.
- Mice are weighed every day for seven consecutive days and % weight loss is calculated in comparison to the initial weight on Day 0. Mice that lose more than 20% of initial body weight or demonstrate shaking or severe systemic responses are euthanized within 24 h.
- Weight loss kinetics of colitis may vary between i.p. and i.v. injection type. In our hands, mice recover weight more quickly after i.p. delivery. However, the route of injection does not alter the timing or severity of colitis development.
- At Days 2-7 post induction, euthanize mice by carbon dioxide asphyxiation according to the approved ethical protocol.
- Collect serum via cardiac puncture for the detection of systemic cytokines levels.
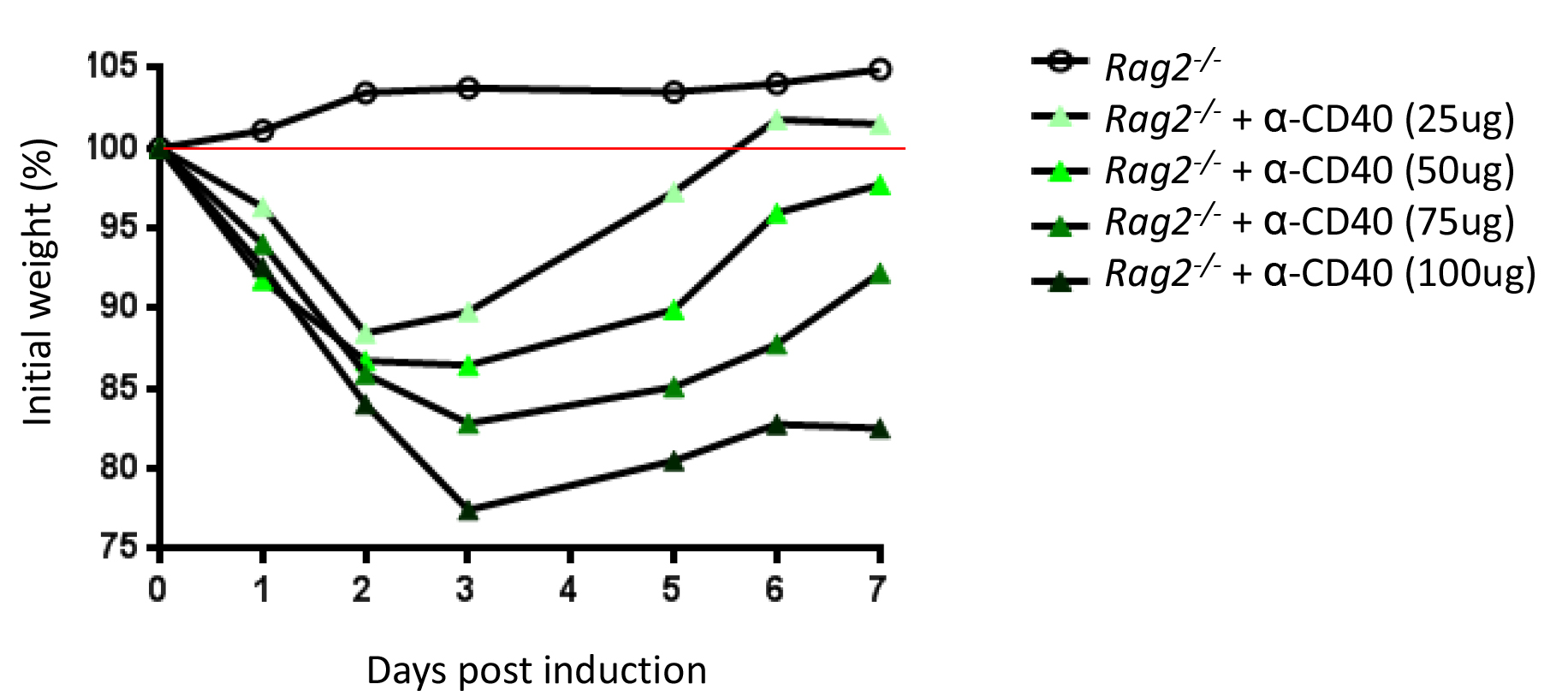
Figure 2. Titration of the anti-CD40 antibody. Rag2-/- mice received a single dose of anti-CD40 antibody i.v. as indicated. Percentage of initial weight over a 7 days period. Initial weight is measured right before injection of anti-CD40 antibody.
- Isolation and preparation of colon tissue/cells for endpoint analysis
- Histopathological analysis by H&E staining
- Harvest colons and remove fecal content by gently dislodging from colon with forceps. Roll colon into a tissue embedding cassette with proximal end in the center and fix in 10% neutral buffered formalin overnight (the cassette needs to be submerged).
- Transfer the cassettes to 70% ethanol, embed in paraffin, section at 4-5 μm and then stain with hematoxylin for 10 min and eosin as previously described (Wang et al., 2017).
- Wash with running water until the water is clear.
- Transfer the slides to the Eosin solution and incubate for 3 min.
- Successively transfer the slides into 70% ethanol for 20 s, 90% ethanol for 20 s, 100% ethanol for 1 min and xylene for 3 min.
- Take out slides from xylene and dry them out in a fume hood.
- Mount and cover the slides.
- Gene expression profiling of the proximal colon
- At the desired time point after induction of colitis, mice are euthanized by carbon dioxide asphyxiation according to approved ethical protocol.
Note: For analysis of genes encoding cytokines produced by the innate immune cells, we recommend euthanizing mice on Days 1 to 3 post induction. For analysis of genes expressed by epithelial cells in response to inflammation, we recommend euthanizing the mice on Day 7 post induction when the disease is maximal. - Harvest a piece of proximal colon (about 0.5 cm long), remove the fecal content by gently dislodging it from colon with forceps, transfer into a sterile polypropylene tube and snap freeze in liquid nitrogen (Figure 3B). Since inflammatory infiltrate can be variable, it is important to cut a vertical section of the colon starting from the base of the cecum and descending into the proximal colon to ensure consistent molecular signature.
Note: To avoid RNA degradation, samples must be snap frozen in liquid nitrogen within 2 min post euthanasia. Frozen proximal colons can be stored at -80 °C for at least 6 months. - Extract RNA and run RT-PCR protocols as described in Bauche et al., 2018.
- At the desired time point after induction of colitis, mice are euthanized by carbon dioxide asphyxiation according to approved ethical protocol.
- Isolation of proximal colon lamina propria cells, ex-vivo stimulation and flow cytometry
Isolation of proximal colon lamina propria cells- At 2-7 days post induction, mice are killed by carbon dioxide asphyxiation according to approved ethical protocol.
- Harvest and collect the proximal colon in a 15 ml Falcon tube filled with 5 ml of 1x HBSS.
- Remove the fat (Figure 3A).
- Transfer the colon into a Petri dish.
- Cut colon open lengthwise and wash by submerging and gently shaking in 50 ml of 1x HBSS until the tissue is clear of fecal matter.
- Cut into 0.5 cm segments.
- Place into a 50 ml Falcon tube with 20 ml of epithelial cell dissociation solution.
- Wrap in parafilm, place sideways in a water bath at 37 °C for 20 min with medium shaking.
- Vortex well for 15 s.
- Filter through a 70 μm strainer, collect and transfer the undigested piece of colon tissues in a new 50 ml Falcon tube.
- Rinse fragments with 20 ml 1x HBSS by gently vortexing for 10 s.
- Transfer colon fragments into a petri dish and chop into tiny pieces with a scalpel.
- Using a 25 ml Serological pipettes, transfer into a new 50 ml tube with 10 ml of enzyme digestion mix then incubate for 20 min in a water bath at 37 °C with shaking as above.
- Vortex for 15 s.
- Collect sup by filtering through a 70 μm cell strainer and put on ice. Pellet cells for 5 min at 375 x g, 4 °C.
- Prepare 100% Percoll solution (9 parts Percoll + 1 part 10x PBS).
- Prepare 40% and 80% Percoll solutions by diluting 100% Percoll with 1x PBS.
- In a 15 ml tube resuspend the pellet in 5 ml of 40% Percoll, using a 5 ml Serological pipettes, and underlay with 5 ml of 80% Percoll for each colon.
- Spin 670 x g for 20 min at 20 °C with no acceleration and no brake.
- Collect interface (white ring) between and the two layers of percoll into a 15 ml tube, wash with 10 ml of complete IMDM media and spin for 5 min at 375 x g, 4 °C.
- Resuspend in 1 ml of complete IMDM and cells are ready to go.
Ex-vivo stimulation- For cytokines analysis, stimulate 2 x 106 viable cells/well in a 96-well plate for 4 h at 37 °C, 5% CO2 with 50 ng/ml PMA, 500 ng/ml Ionomycin and Brefeldin A (1/1,000) in 200 μl of complete IMDM.
- Spin for 5 min at 375 x g, 4 °C and proceed to flow cytometry staining.
Flow cytometry staining- Incubate cells in 100 μl of 1x PBS with 0.3 μl of live/dead fixable dead cell stains and 5 μl of mouse BD Fc block per well for 25 min at 4 °C in the dark.
- Wash with 100 μl of 1x PBS per well and spin for 5 min at 375 x g.
- Incubate cell suspension with 0.2 μg of anti-mouse conjugated antibodies (i.e., CD45, CD3, CD4) in 100 μl of BD stain buffer per well for 25 min at 4 °C in the dark.
- Wash with 100 μl of BD stain buffer and spin for 5 min at 375 x g.
- Resuspend cell suspension in 100 μl of 1x Fix/perm buffer (BD) and incubate for 45 min at 4 °C in the dark.
- Wash with 1x Perm/Wash buffer and spin for 5 min at 375 x g.
- Incubate cell suspension with 0.2 μg of anti-mouse conjugated antibodies (i.e., RORγt, IL-22) in 100 μl of 1x Perm/Wash buffer per well for 45 min at 4 °C in the dark.
- Wash with 1x Perm/Wash buffer and spin for 5 min at 375 x g.
- Resuspend cells in 300 μl of 1x Perm/Wash buffer. Analyze and acquire data using a flow cytometer (we used LSRII flow cytometer from BD and analyzed data on FlowJo). Samples can be stored overnight at 4 °C in the dark.
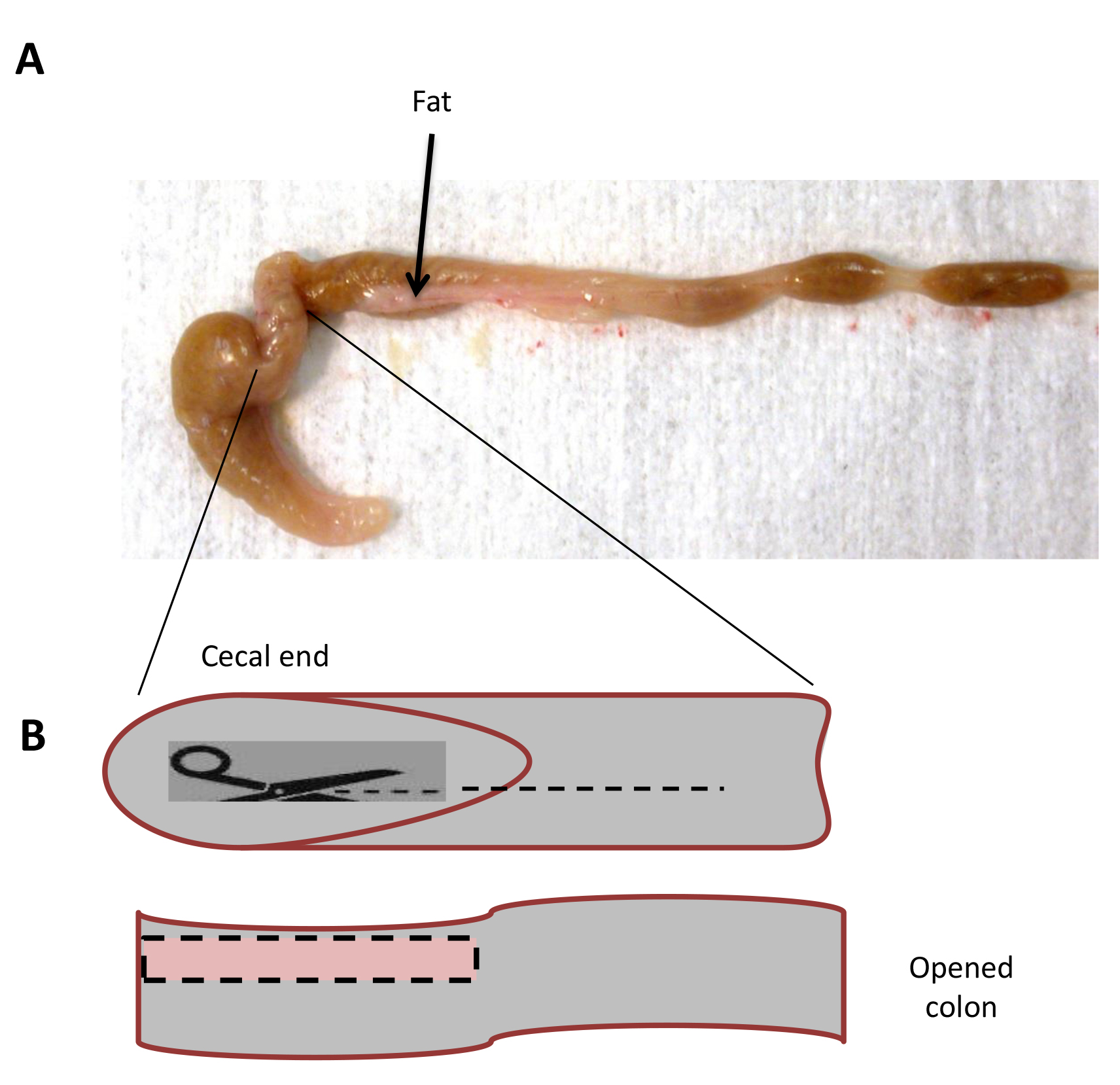
Figure 3. Preparation of colon tissue. A. Attached fat tissue (here shown by arrows) must be removed from the colon as fat can decrease the viability of isolated colon lamina propria. B. Taqman Sampling: The colon is separated from the cecum and carefully dissected from host by removing all the fat and connective tissue. The colon is cut from the proximal (cecal) end, down the midline and until the distal end. Flatten out the tissue, cut a 2-3 cm strip (pink square) and immediately snap freeze using polypropylene tubes in liquid nitrogen.
- Histopathological analysis by H&E staining
Data analysis
- Tissues are scored for severity of disease by a pathologist according to three criteria (each criterion is scored from 0 to 3, 0 = negative, 1 = mild, 2 = moderate and 3 = severe):
Inflammation: when present is characterized by infiltration of large numbers (60%-70%) of mononuclear cells (macrophages and lymphocytes) and 30%-40% of neutrophils and band cells. The scoring of inflammation includes severity of infiltration, loss of glands, erosion, and dilatation of glandular lumina, presence of crypt abscess and degeneration of epithelial cells.
Apoptosis: The prevalence of apoptotic bodies is scored on a scale of 0-3: 0 = negative; 1 = low; 2 = moderate; 3 = high.
Regeneration: Regenerative changes assessed include scoring the prevalence of mitotic figures in the upper 1/3 of the mucosa, nuclear density (nuclear crowding) within individual glandular structures, regularity of the surface epithelium. Regeneration is scored on a scale of 0-3: 0 = negative; 1 = low; 2 = moderate; 3 = high. - For gene expression profiling, average the normalized values from isotypes
Calculate the fold change over the isotype control using the following formula:
fold change = (Normalized value of the sample)/(Average normalized value of the isotype control group) - For flow cytometry analysis, acquire at least 5 x 105 colon lamina propria cells
- The events are gated on FSC-A and SSC-A, then gated on FSC-A and FSC-H to remove doublets.
- Exclude cellular debris and dead cells (using LIVE/DEAD fixable stain).
- To study the phenotype of infiltrating immune cells, gate on CD45+ cells.
- For analysis of IL-22-producing ILC3, gate on live cells, lineage (CD11c, CD11b, NK1.1)–, CD90.2high, CD45int, RORγt+, IL-22+ cells. For further details, gating strategy can be found in previous publication (Bauche et al., 2018).
- Statistical analysis was performed using Prism 7. Unpaired t-test is used as statistical test. A P value < 0.05 is considered statistically significant.
Recipes
- Epithelial cell dissociation solution
1x HBSS
Penicillin/streptomycin (1,000 U/ml)
5 mM EDTA
10 mM HEPES - Enzyme digestion mix
1x HBSS
Penicillin/streptomycin (1,000 U/ml)
10% FBS
2 mg/ml collagenase type VIII
30 U/ml of DNase I
50 U/ml of Dispase
Alternately, 1 mg/ml of collagenase D (Sigma) can be used to preserve cell surface molecules when analyzing by FACS - Complete IMDM media, 500 ml
450 ml of IMDM, GlutaMAX supplement
1,000 U/ml of Penicillin/streptomycin
1 mM of Sodium pyruvate
0.55 mM of 2-Mercaptoethanol
50 ml of Heat inactivated FBS
Acknowledgments
We would like to thank Gil Asio and Danye Cheng for processing colon samples. We appreciate Lakshmanan Annamalai and Jennifer H. Yearley for their expertise in analyzing and scoring the histological samples. Wendy Blumenschein and Jeff Grein for running gene expression profile.
Competing interests
The authors declare no competing financial interests.
Ethics
Mice were maintained under specific pathogen-free conditions and kept in microisolators with filtered water at Merck Research Laboratories (MRL) animal facility, Palo Alto. All animal procedures were approved by the Institutional Animal Care and Use Committee of MRL in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC).
References
- Bauche, D., Joyce-Shaikh, B., Jain, R., Grein, J., Ku, K. S., Blumenschein, W. M., Ganal-Vonarburg, S. C., Wilson, D. C., McClanahan, T. K., Malefyt, R. W., Macpherson, A. J., Annamalai, L., Yearley, J. H. and Cua, D. J. (2018). LAG3+ regulatory t cells restrain interleukin-23-producing cx3cr1+ gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity 49(2): 342-352: e345.
- Cayatte, C., Joyce-Shaikh, B., Vega, F., Boniface, K., Grein, J., Murphy, E., Blumenschein, W. M., Chen, S., Malinao, M. C., Basham, B., Pierce, R. H., Bowman, E. P., McKenzie, B. S., Elson, C. O., Faubion, W. A., Malefyt Rde, W., Kastelein, R. A., Cua, D., McClanahan, T. K. and Beaumont, M. (2012). Biomarkers of therapeutic response in the il-23 pathway in inflammatory bowel disease. Clin Transl Gastroenterol 3: e10.
- Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., Panaccione, R., Ghosh, S., Wu, J. C. Y., Chan, F. K. L., Sung, J. J. Y. and Kaplan, G. G. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390(10114): 2769-2778.
- Uhlig, H. H., McKenzie, B. S., Hue, S., Thompson, C., Joyce-Shaikh, B., Stepankova, R., Robinson, N., Buonocore, S., Tlaskalova-Hogenova, H., Cua, D. J. and Powrie, F. (2006). Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity 25(2): 309-318.
- Wang, C., Yue, F. and Kuang, S. (2017). Muscle histology characterization using H&E staining and muscle fiber type classification using immunofluorescence staining. Bio-protocol 7(10): e2279.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Joyce-Shaikh, B., Cua, D. J. and Bauché, D. (2019). Induction and Analysis of Anti-CD40-induced Colitis in Mice. Bio-protocol 9(3): e3153. DOI: 10.21769/BioProtoc.3153.
Category
Immunology > Animal model > Mouse
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









