- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunohistochemical Staining of TLR4 in Human Skeletal Muscle Samples
Published: Vol 9, Iss 2, Jan 20, 2019 DOI: 10.21769/BioProtoc.3144 Views: 6565
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells
Harsimranjit Sekhon [...] Stewart N. Loh
Mar 20, 2025 2262 Views

Dissection and Whole-Mount Immunofluorescent Staining of Mouse Hind Paw Muscles for Neuromuscular Junction Analysis
Rebecca L. Simkin [...] James N. Sleigh
May 20, 2025 3909 Views

Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2374 Views
Abstract
Growing evidence suggests the involvement of TLR4, a receptor in the innate immune system, in muscle loss in uremia. Recently, we have evaluated TLR4 in human skeletal muscle from chronic kidney disease patients, by immunohistochemistry and image analysis. Unlike the commonly-used Western blot method, immunohistochemistry allows for the observation of protein distribution in the intact tissue while, image analysis, its quantification. In fact, our data highlighted our hypothesis that an enhanced TLR4 skeletal muscle cell expression contributes to the activation of the downward inflammatory pathway in uremic sarcopenia. In this protocol, we describe the procedure for immunostaining TLR4 in human skeletal muscle and for quantifying it by image analysis.
Keywords: Human skeletal muscleBackground
TLRs are a family of receptors in the innate immune system that play crucial roles in the early defense stage against foreign agents (Aderem and Ulevitch, 2000; Takeda and Akira, 2001; Kawai and Akira, 2010). TLR4 recognizes pathogen-associated or endogenous antigens with common molecular patterns, including debris from apoptotic and necrotic cells (Mkaddem et al., 2010), heat shock proteins (Andersson et al., 2018) and oligosaccharides (Barrera et al., 2015). In addition, the activation of TLRs is an important link between innate and adaptive immune systems (Takeda and Akira, 2001). An emerging hypothesis underpins the role of innate immunity in the development and progression of uremic cachexia as observed in cancer (Zhang et al., 2017) and cardiac cachexia (Avlas et al., 2015) and there is also growing recognition that uremia stimulates inflammation, thus regulating the pathways leading to muscle catabolism (Jankowska et al., 2017). Among TLRs, TLR4 is involved in muscle loss, by the engagement of several ligands which induce and amplify the inflammatory response through the activation of the transcription factor NF-κB (Takeda and Akira, 2001).
Recently, we have demonstrated, both ex vivo and in vitro, that in chronic kidney disease (CKD) uremia promotes muscle inflammation through an up-regulation of TLR4. In turn, it activates downward inflammatory signals, such as TNF-α and NF-κB-regulated genes (Verzola et al., 2017). In human skeletal muscle, we have analyzed TLR4 expression by immunohistochemistry and Western blot, the most used approaches for protein analysis (Verzola et al., 2017). As far as we are aware, most of scientific reports written in the last 5 years, available on https://www.ncbi.nlm.nih.gov/pubmed, mainly evaluate skeletal muscle TLR4 expression by Western blot, that determines its relative abundance within a muscle sample, but does not allow for the mapping of the distribution among the various fibers whereas Immunohistochemistry provides a “picture” and enables the observation of the protein in the intact tissue. However, since both methods have advantages and disadvantages, a combination of the two approaches is required to obtain a correct and complete characterization of TLR4 expression. In addition, the data obtained by immunohistochemistry on TLR4 protein expression, also provides details concerning its amount and distribution along the muscular fibers and the cellular components, supporting the hypothesis that an enhanced expression of TLR4 contributes to the upregulation of native immunity in CKD skeletal muscle cells. Furthermore, thanks to the computer-assisted image analysis and the resulting quantification of the intensity of staining, it was possible firstly, to differentiate between the 2 groups, the healthy patients and the CKD patients, and secondly to statistically evaluate each of them.
Materials and Reagents
- Pipette tips (StarLab, catalog numbers: S1111-3700 [10 µl], S1113-1700 [200 µl], S1111-6700 [1,000 µl])
- Aluminum foil (CA.DIS., catalog number: 50002.24)
- Petri dishes 60 mm (Euroclone, catalog number: ET2060)
- 50 ml conical tubes (Eppendorf, catalog number: E162071N)
- 15 ml conical tubes (Eppendorf, catalog number: E160926K)
- Disposable gloves (WRP, catalog number: D1502-18)
- S35 Feather disposable microtome blades (PFM Medica AG, catalog number: 02OK07500000)
- Adhesion microscope slides (Laboindustria S.p.A., catalog number: 33820)
- Coverslips for optical microscopy 24 x 50 x 0.16 mm (DiaPath, catalog number: 061051)
- Pap pen for immunostaining (Merck group, catalog number: Z377821-1EA)
- Paper towel (Paperdi, catalog number: RL1M180)
- Small brushes
- Vacuum Filter System 500 ml 0.45 μm (Euroclone, catalog number: EPVPE45500)
- DPBS (Euroclone, catalog number: ECB400L)
- Ethanol (ITW Reagents, catalog number: 141086)
- Xylene mix of isomers (Carlo Erba Reagents, catalog number: 392603)
- Paraplast Bulk (Tissue embedding medium) (Leica Biosystem, catalog number: 39602012)
- Eukitt (Fluka Analytical, catalog number: 03989)
- Deionized water
- Distilled water
- Methanol (Carlo Erba Reagents, catalog number: 414814)
- Hydrogen peroxide 30% in water (Merck, catalog number: 216763-M)
- Hydrochloric acid, 37% (Merck, catalog number: 30721-M)
- Bovine serum albumin (Merck, catalog number: A3912)
- Avidin/Biotin Blocking kit (Vector Laboratories, catalog number: SP-2001)
- Monoclonal Antibody to TLR4 (Abcam, catalog number: 76B357.1)
- Mouse IgG (Vector Laboratories, catalog number: I-2000)
- Biotinylated anti-mouse IgG (H+L) affinity purified (VectorLaboratories,catalog number: BA-2000)
- Streptavidin/HRP (Dako, catalog number: P0397)
- DAB (3,3’-diaminobenzidine tetrahydrochloride) Enhanced Liquid Substrate System (Merck, catalog number: D3939)
- Carazzi’s Hematoxylin (Bio-Optica, catalog number: W01030708)
- Paraformaldehyde powder EM grade (Polysciences, catalog number: 0380)
- Potassium chloride (KCl) (Carlo Erba Reagents, catalog number: 471177)
- Potassium phosphate monobasic (KH2PO4) (Carlo Erba Reagents, catalog number: 471687)
- Di-sodium hydrogen phosphate anhydrous (Na2HPO4) (Carlo Erba Reagents, catalog number: 480141)
- Sodium chloride (NaCl) (Carlo Erba Reagents, catalog number: 368257)
- Citric acid (CARLO ERBA Reagents, catalog number: 403727)
- Tri-sodium citrate dihydrate (Carlo Erba Reagents, catalog number: 479487)
- Sodium azide (NaN3) (Merck, catalog number: S-2002)
- 2% paraformaldehyde (pH 7.4) (see Recipes)
- PBS (for diluting secondary antibodies, streptavidin/HRP and washing and dissolving BSA) (see Recipes)
- Unmasking buffer (10 mM sodium citrate buffer, pH 6) (see Recipes)
- Buffer for diluting primary antibody (see Recipes)
- Peroxidase inhibition solution (see Recipes)
- Blocking solution (5% BSA) (see Recipes)
Equipment
- Tweezers (Thermo Fisher Scientific, catalog number: 10303611)
- Forceps (Exacta Optech, catalog number: 9204222)
- Coverglass forceps (Leica Biosystems, catalog number: 38DI26286)
- Sterile mono-use scalpel
- Beakers
- Glass-staining dishes with lid (Vetrotecnica, catalog number: 01406000)
- PMP vertical staining jars with lids (Vetrotecnica, catalog number: 06035500)
- Pipettes (0.5-10, 10-100 and 100-1,000 μl, Eppendorf, catalog numbers: 3111000.122, 3111000.149, 3111000.165)
- Leica Reichert-Jung 2040 autocut Microtome
- Histology Bath (Falc Instruments, catalog number: 610.1030.00)
- Metal base molds 24 x 24 mm (Leica, catalog number: 38013082)
- Tissue Embedding Rings (Biosigma, catalog number: 040272)
- Drying oven/bench top (Melag)
- Humidity chamber (Merck, catalog number: H6644)
- Light microscope (Leica, model: Leica DMLB, catalog number: 020-519.502)
- High-resolution digital camera (Leica, model: Leica DC 300F, catalog number: 12447115)
- Heating magnetic stirrer (Velp Scientifica, catalog number: F20500413)
- Fume cupboard (Bicasa, catalog number: Logica 1.2) Microwave oven Saturno (MG821ABH) (DPE Elettrodomestici, catalog number: 0277)
Software
- Image analysis software (Leica QWin) (Leica, Cambridge, UK)
- GraphPad Prism® (version 5.02) (GraphPad Software San Diego, CA, U.S.A.)
Procedure
- Sample collection
- Preparation (1)
- Wrap clean tweezers and forceps in an aluminum foil and autoclave them.
- Prepare two 60 mm dishes per muscle sample, pipette 3 ml cold (~5 °C) sterile DPBS into each dish.
- Collection of human muscle biopsies from the rectus abdominis muscle
Notes:- Chronic Kidney Disease (CKD) muscle biopsies were obtained during peritoneal dialysis catheter insertion procedures.
- Control biopsies were taken from otherwise healthy subjects during elective surgery for abdominal wall hernias.
- Informed consent was obtained from all subjects.
- Preparation (2)
- Give muscle tissue a code, that identifies each specimen for each patient.
- Transfer the muscle biopsies in a 60 mm dish containing 3 ml cold (~5 °C) DPBS.
- Clean tissue muscle by tweezers and a sterile mono-use scalpel from any visible connective and adipose tissue.
- Reduce the specimen size with a sterile mono-use scalpel, if necessary. Generally 0.4 x 0.4 cm is enough.
- Wash the muscle in another 60 mm dish containing 3 ml of cold (~5 °C) DPBS.
- Preparation (1)
- Paraffin slides preparation
- Muscle processing
- Transfer the tissue into a 15 ml conical tube containing 10 ml of 2% paraformaldehyde and fix overnight at 4 °C.
- Discard 2% paraformaldehyde.
- Dehydrate the tissue by passing it through a series of solutions of increasing Ethanol concentration:
10 ml 50% ethanol for 60 min at room temperature
10 ml 70% ethanol for 60 min at room temperature
10 ml of 100% ethanol for 60 min at room temperature
10 ml of 100% ethanol overnight at 4 °C - Clear with xylene for 4 h at room temperature.
- Discard the xylene.
- Infiltrate the muscle tissue in melted paraffin at 56 °C for 60 min.
- Embedding
- Use forceps to transfer the muscle tissue to the well of the embedding base mold.
- Snap the base of the tissue cassette into the embedding base mold.
- Fill the mold with melted paraffin.
- Previously the embedding base mold is cooled down on a block of ice, and each step is carried out as quickly as possible!
- Allow the paraffin to solidify at -20 °C.
- Sectioning
- Heat the deionized water up to 56 °C in a histology bath.
- Mount the paraffin block on the microtome.
- Write down the code specimen on the slide with a pencil.
- Cut thin sections (5 µm).
- Pick up the sections using small brushes and transfer them into the histology bath.
- Dip a microscope slide into the histology bath, allowing the tissue section to adhere to its surface.
- Dry the slides in an oven at 56 °C from 1 h up to overnight.
- Muscle processing
- Immunohistochemistry staining
Notes:- Before staining, the paraffin must be removed.
- All steps must be performed in the fume cupboard.
- De-paraffinization/Rehydration
- Prepare 4 glass staining dishes, each with a lid, containing:
Ethanol 50%
Ethanol 70%
Ethanol 100%
Xylene - Transfer the slides into:
Xylene for 10 min Ethanol 100% for 10 min Ethanol 70% for 10 min Ethanol 50% for 10 min Deionized water for 1 min
- Prepare 4 glass staining dishes, each with a lid, containing:
- Antigen Unmasking
- Transfer the slides into the PMP vertical staining jar with lid.
- Heat slides in the microwave submersed in 60 ml 10 mM sodium citrate buffer and partially covered with lid until boiling is initiated.
- Sub-boil for 15 min.
- Cool slides on bench top for 20 min.
- Staining
- Wash the slides in deionized water for 30 s.
- Block endogen peroxidases with 3% hydrogen peroxide (peroxidase inhibition solution) for 15 min (repeat twice).
Note: Prepare freshly 3% hydrogen peroxide at the time of the use and for each change time. - Wash the slides in deionized water for 30 s.
- Wash the sections in PBS for 5 min.
Note: From this point forward, transfer the slides in the humidity chamber. Wash the slides in the glass staining dish. - Block each section with 100-200 µl of 5% BSA for 20 min, at room temperature.
- Wash the sections in PBS for 5 min for 3 times.
- Draw a circle around the tissue section by a pap pen.
- Block each section with a drop of avidin for 15 min at room temperature.
- Repeat Step C3f.OK
- Block each section with a drop of biotin for 15 min, at room temperature.
- Repeat Step C3f.
- Dilute the primary antibodies in 1% BSA-PBS.
Note: The bottom of the humidity chamber was covered with water. For primary antibody dilution, we consulted product datasheet. According to TLR4, 1:100, the negative control was obtained incubating the section with mouse IgG in the same conditions. - Incubate each section with 100 µl overnight at room temperature.
- See Repeat Step C3f.
- Dilute the Biotinylated Anti-Mouse IgG in PBS in the ratio of 1:300.
- Incubate each section with 100 µl, 30 min at room temperature.
- Repeat Step C3f.
- Dilute Streptavidin/HRP in PBS in the ratio of 1:500.
- Repeat Step C3p.
Note: During Steps C3m, C3p and C3s, ensure that the slides dry up. - Repeat C3f.
- Prepare DAB (3,3’-diaminobenzidine tetra hydrochloride) Enhanced Liquid Substrate System according to data sheet.
- Incubate each section with 100 µl DAB solution for 15 min at room temperature.
Note: Monitor the staining development under a microscope. - Repeat Step C3f.
Note: Wash the slides in a glass staining dish. - Immerse slides in deionized water.
- Counterstain the sections with Carazzi’s Hematoxylin for 1 min.
- Rinse under tap water for 5 min.
- Transfer the slides into the deionized water.
- Rehydration and the mounting of the sections
Transfer the slides into:
Note: This second set of ethanol solutions and xylene must be different from those used above for de-paraffinization.Ethanol 50% for 2 min Ethanol 70% for 2 min Ethanol 100% for 2 min Xylene for 2 min - Preparing permanently mounted sections
- Place 2-3 drops of Eukitt over the section.
- Avoid trapping air bubbles, lowering the coverslip carefully from one side of the droplet by the coverglass forceps.
Note: There should be enough mounting medium to completely cover the bottom of the cover slide. Excess Eukitt can be removed by a paper towel (Figure 1). - Leave slides in the fume cupboard for 24 h.
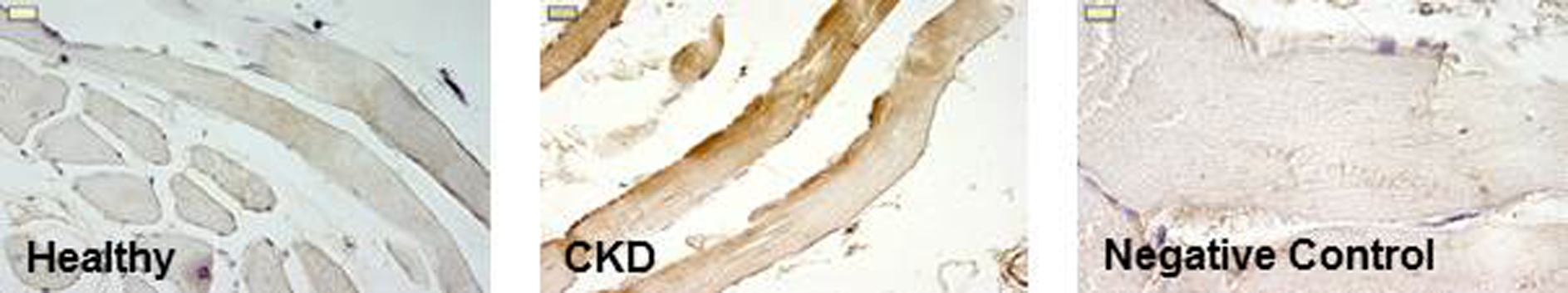
Figure 1. TLR4 staining in Healthy and CKD muscle. Negative control is obtained by incubating the slide with the same first antibody isotype (Magnification400-1,000x; Scale bar = 10 µm).
Data analysis
Image analysis
In order to quantify staining differences, among the muscle specimens, we used image analysis. It is a powerful tool that provides meaningful quantitative data using analysis software that segments pixels in a digital image based on features such as color (i.e., RGB). Image analysis was performed by Leica QWin Image Analysis System (Leica, Cambridge, UK). The video image is generated by a CCD camera connected through a frame grabber to a PC. Single images are digitized for image analysis at 256 gray levels. Imported data are analyzed quantitatively by QWin (Leica, Cambridge, UK) and statistical analysis was performed by GraphPad Prism®.
- Choose the best magnification to analyze immunostained muscle and maintain the same for each specimen and analysis. We used 400x magnification. Adjust exposure time and gain setting (Figure 2).
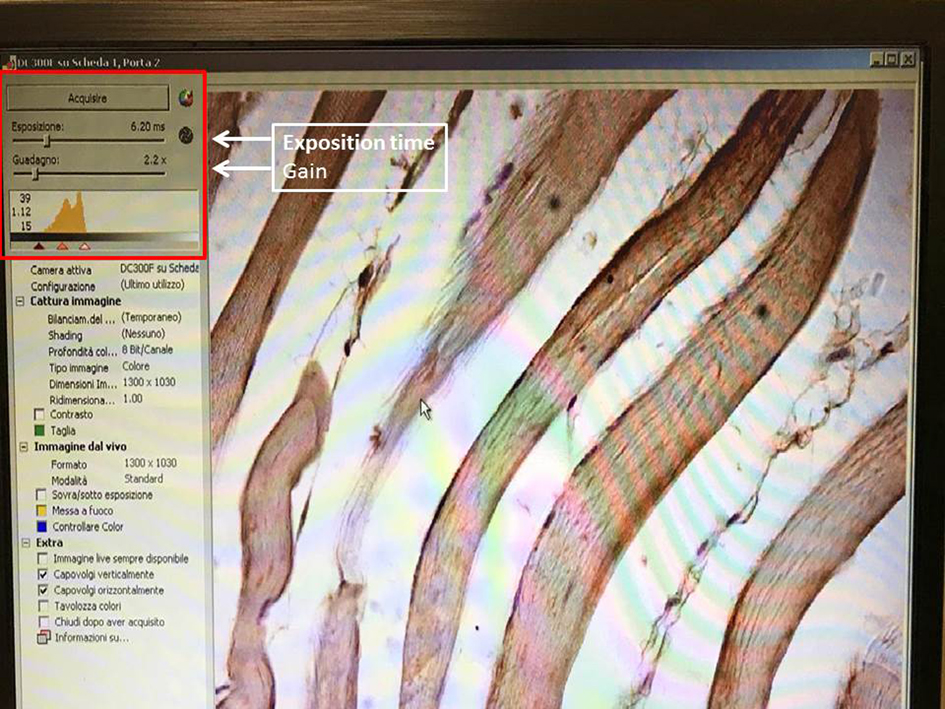
Figure 2. Exposure time and gain setting. The screenshot shows the exposure time and gain setting permit the creation of the best conditions for image acquisition. - Set the white color balance, selecting a little small frame on the slide using the mouse (Figure 3).
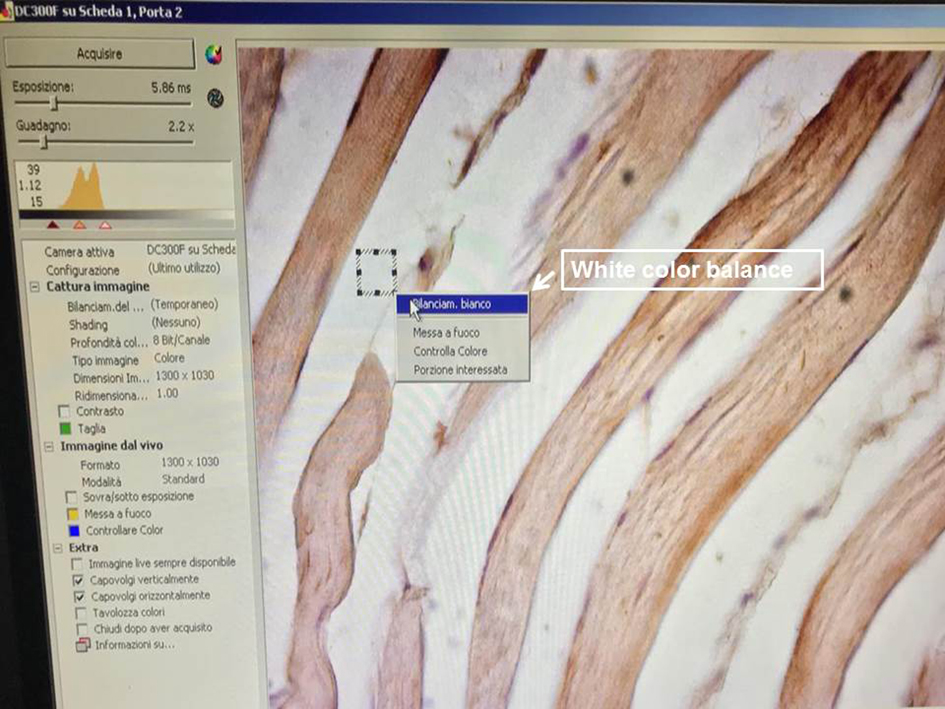
Figure 3. White color balance setting. This function performs a color correction in order to well balance the color image. It is one of the most important settings for the acquisition. For the white balance, click using the left button of the mouse on one corner of the white area in order to form a small frame. As soon as you release the button, a context menu appears containing the white balance command as shown in the photo and underlined by a white arrow. - Adjust fine focus, position and light level (Figure 4).
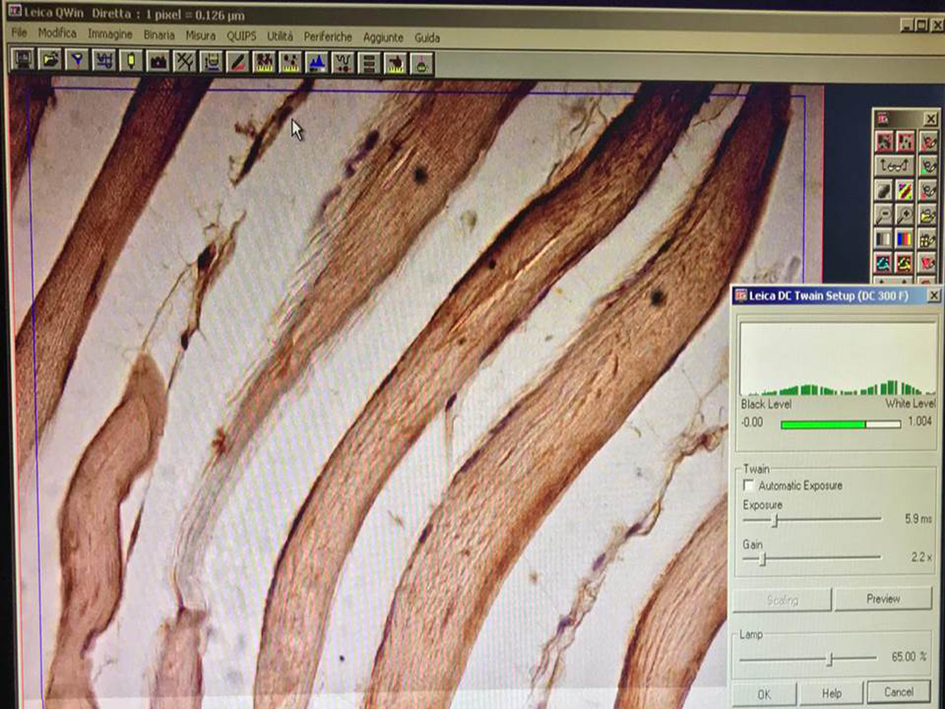
Figure 4. Adjusting fine focus, position and light level. The histogram on the right indicates the extent to which the image is under-exposed or over-exposed and also assists in adjusting the lighting condition. Optimal exposure is indicated by the green histogram.
Note: Optimal image exposure is critical for nearly all computer vision applications. When an image is acquired, it is important to find a compromise between exposure time and image brightness. If the image is under- or over-exposed, features or the texture are lost, colors washed out, and the overall perceptual quality of the image is decreased. - Acquire the image.
- Set red, blue and green peaks in order to mark only brown-stained area and exclude negative areas (Figure 5).
Note: Maintain Ensure the setting for each specimen and analysis is maintained.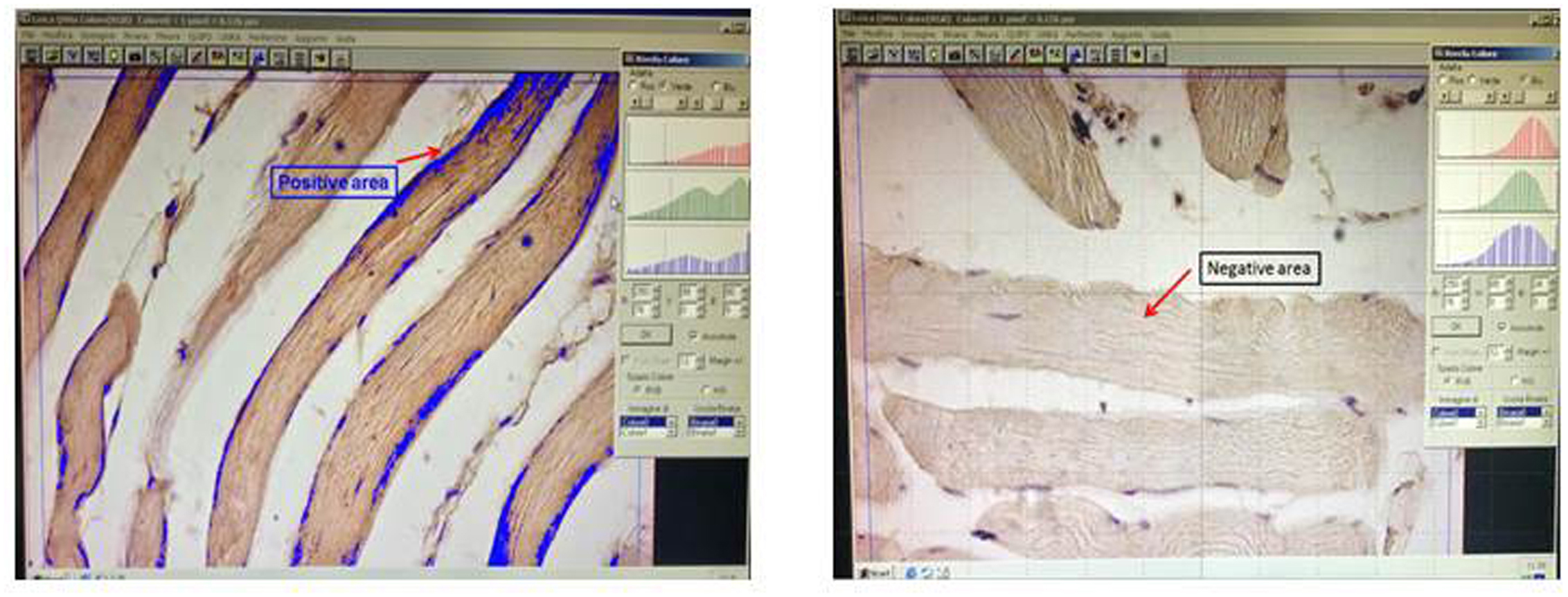
Figure 5. Setting red, blue and green peaks. The histograms on the right are set up in order to distinguish between stained and unstained areas. Positive signals are shown in blue. - Select a single fiber for each field and measure it (Figure 6).
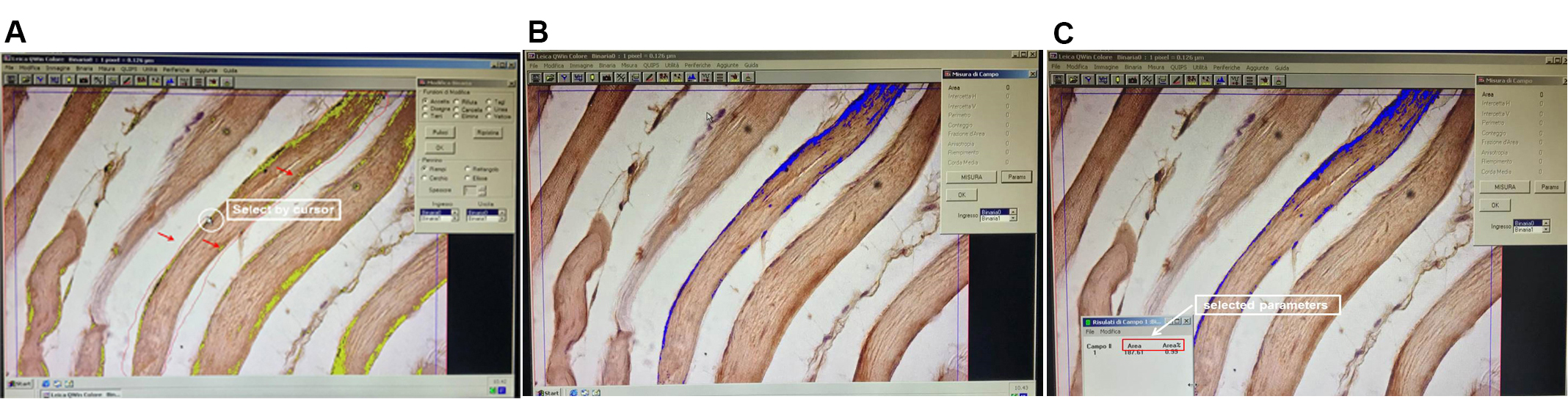
Figure 6. Selection and measurement of single fiber. It is advisable to measure one fiber at a time because the number of muscle fibers is different in each field. A. With cursor (highlighted by the white circle), select one fiber (the fifth from your left) as shown by the thin red line indicated by the three red arrows. B. Only the selected fiber is evaluated as indicated in blue. C. Dialog box on the left displays the values of selected parameters. - Repeat measuring of at least 20-25 fibers (Figure 7).
Note: The fibers should be chosen randomly, evaluating both the negative and the positive ones.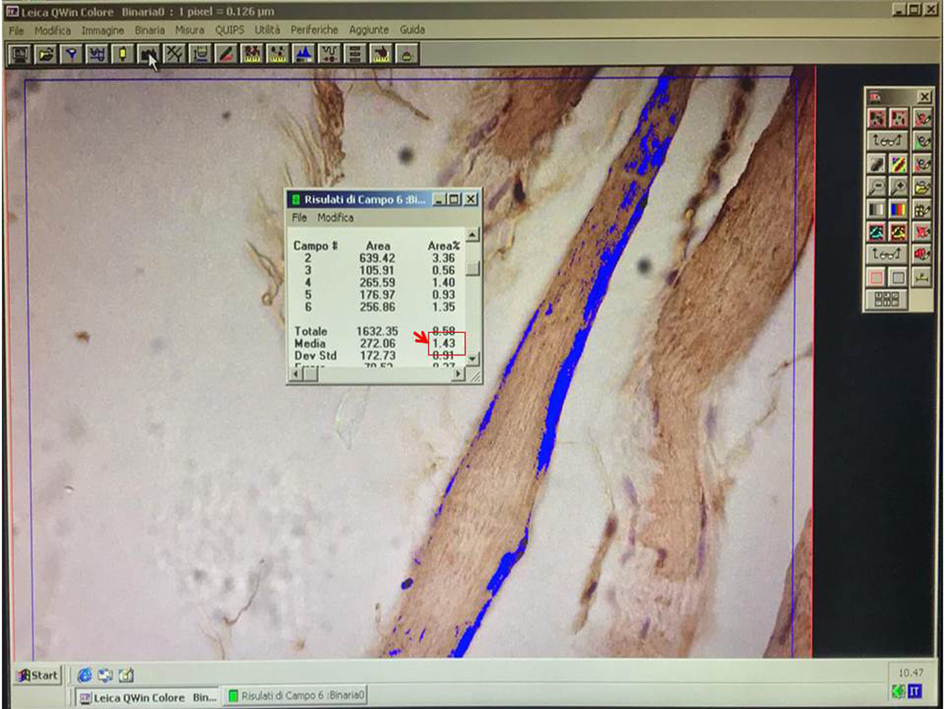
Figure 7. Example of field measurement. In the dialog box, an example of field measurements is reported. The red square highlights the average value. - When the analysis of the specimen is completed, the average of AREA % is considered.
All measures are reported in an Excel sheet as shown below in Figure 8.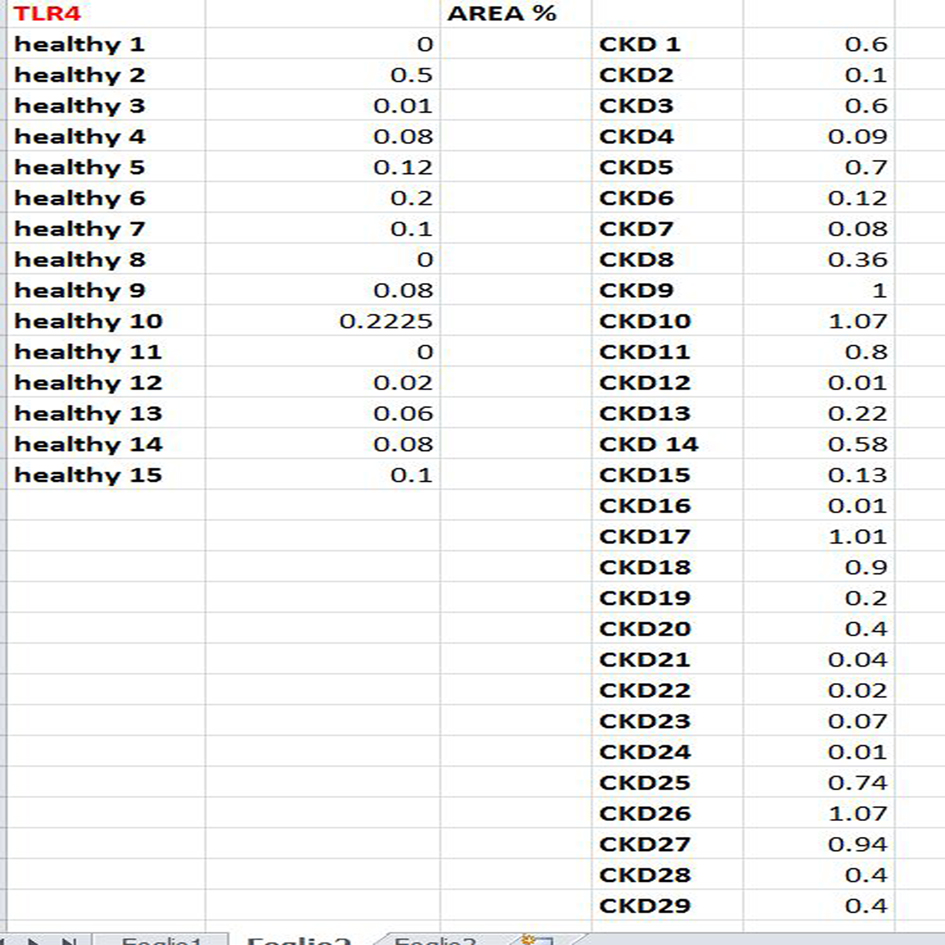
Figure 8. Spreadsheet of TLR4 measures - At this point, the data is ready to be submitted for statistical analysis performed by GraphPad Prism® (Figure 9).
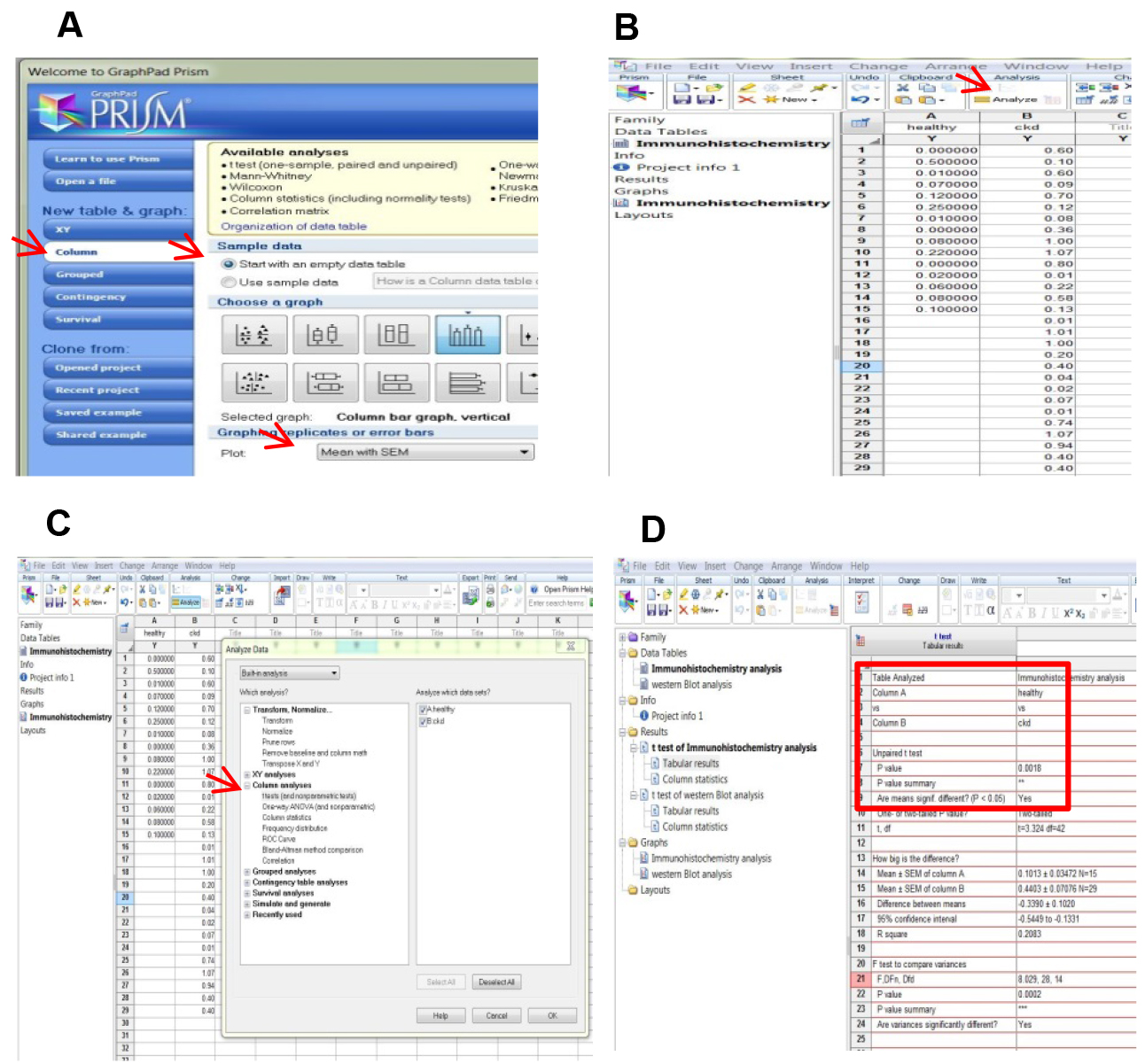
Figure 9. Statistical analysis by GraphPad Prism®. A. Open GraphPad Prism® and select the parameters indicated by the red arrows. B. Start from data table and click analyze as indicated by the red arrow. C. Choose the analysis on the left side of the dialog box. In this case, we choose t-test which measures a variable (the mean of intensity staining) in two groups assuming that the values follow a Gaussian distribution. D. The difference between the mean of the two groups is statistically significant as suggested by the P value. - Comparison between the TLR4 evaluation by immunohistochemistry and Western Blot (WB)
We have randomly chosen muscle samples from 5 healthy and 12 CKD subjects, and the extracted proteins were separated by WB. The intensity of the TLR4 expression was evaluated as described in Verzola et al. (2017). Also by WB, the difference between the two groups is statistically significant (Figure 10).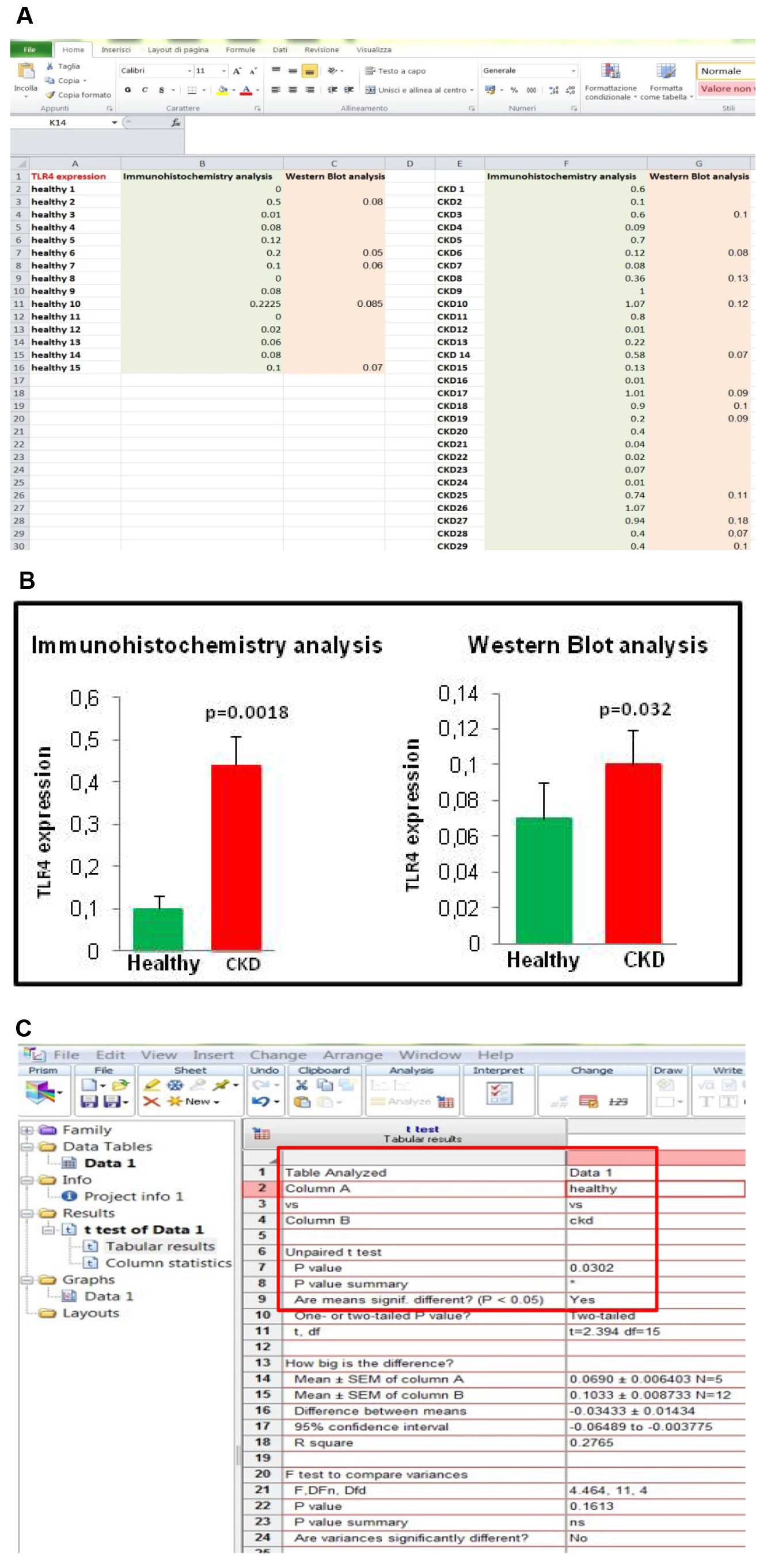
Figure 10. Comparison between TLR4 evaluation by immunohistochemistry and WB. A. Spreadsheet of TLR4 measurements by the two methods. B. Graphs show that TLR4 is increased in CKD muscle by using both methods. C. Also by WB, the difference between the mean of TLR4 of the two groups is statistically significant as suggested by the P value.
Recipes
- Paraformaldehyde 2%, pH 7.4 (store at 4 °C for up to one month)
DPBS (Euroclone)
Paraformaldehyde powder EM grade
For 500 ml of 2% paraformaldehyde, in a glass beaker:- Mix 10 g paraformaldehyde with 400 ml DPBS on a stir plate in a ventilated hood
- Heat the solution, while stirring, to approximately 60 °C
Note: Be careful that the solution does not boil. - Once the paraformaldehyde is dissolved, the solution is cooled and filtered
- Adjust the volume of the solution to 500 ml with DPBS
- PBS (It can be stored at room temperature) (500 ml) (for washing and diluting biotinylated anti-mouse antibody and Streptavidin/HRP)
0.2 g KCl
0.2 g KH2PO4
1.08 g Na2HPO4 8 g NaCl
In a beaker:- Dissolve the salts in 400 ml of deionized water on a stir plate
- Add some drops of HCl 37% for adjusting the pH to 7.4
- Adjust the volume of the solution to 500 ml with deionized water
- Unmasking buffer (10 mM sodium citrate buffer) (100 ml)
- Solution A (100 ml)
2.1 g citric acid
100 ml distilled water - Solution B (100 ml)
2.94 g tri-sodium citrate dihydrate
100 ml distilled water
At the time of use, mix 2.25 ml Solution A + 10.25 ml Solution B + 125 ml distilled water
Check pH = 6 - Solution A (100 ml)
- Buffer for diluting primary antibody
100 ml PBS
1 g BSA
0.1 g NaN3- In a beaker, dissolve BSA and NaN3 in 80 ml of PBS at 4 °C
- Adjust the volume of the solution to 100 ml with PBS
- Store at 4-8 °C up to one month
- Peroxidase inhibition solution
90 ml methanol
10 ml 30% hydrogen peroxide
Note: Prepare freshly at the time of use. - Blocking solution (5% BSA)
50 ml PBS
2.5 g BSA
Store at 4 °C for 1 week
Acknowledgments
This study was supported by grants from the “Ministero dell’Università e della Ricerca Scientifica eTecnologica” and by from the University of Genoa (Cofinanziamento di Ateneo).
Competing interests
Authors have no conflicts of interest or competing interests to disclose.
Ethics
The study was approved by the Ethical Committee of the Department of Internal Medicine (Di.M.I.) at the University of Genoa. All subjects were informed of the nature, purposes, procedures, and possible risks of the study before their informed consent was obtained.
References
- Aderem, A. and Ulevitch, R. J. (2000). Toll-like receptors in the induction of the innate immune response. Nature 406(6797): 782-787.
- Andersson, U., Yang, H. and Harris, H. (2018). High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin Immunol 38: 40-48.
- Avlas, O., Bragg, A., Fuks, A., Nicholson, J. D., Farkash, A., Porat, E., Aravot, D., Levy-Drummer, R. S., Cohen, C., Shainberg, A., Arad, M. and Hochhauser, E. (2015). TLR4 expression is associated with left ventricular dysfunction in patients undergoing coronary artery bypass surgery. PLoS One 10(6): e0120175.
- Barrera, M. J., Aguilera, S., Veerman, E., Quest, A. F., Diaz-Jimenez, D., Urzua, U., Cortes, J., Gonzalez, S., Castro, I., Molina, C., Bahamondes, V., Leyton, C., Hermoso, M. A. and Gonzalez, M. J. (2015). Salivary mucins induce a Toll-like receptor 4-mediated pro-inflammatory response in human submandibular salivary cells: are mucins involved in Sjogren's syndrome? Rheumatology (Oxford) 54(8): 1518-1527.
- Jankowska, M., Cobo, G., Lindholm, B. and Stenvinkel, P. (2017). Inflammation and protein-energy wasting in the uremic milieu. Contrib Nephrol 191: 58-71.
- Kawai, T. and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11(5): 373-384.
- Mkaddem, S. B., Bens, M. and Vandewalle, A. (2010). Differential activation of Toll-like receptor-mediated apoptosis induced by hypoxia. Oncotarget 1(8): 741-750.
- Takeda, K. and Akira, S. (2001). Regulation of innate immune responses by Toll-like receptors. Jpn J Infect Dis 54(6): 209-219.
- Verzola, D., Bonanni, A., Sofia, A., Montecucco, F., D'Amato, E., Cademartori, V., Parodi, E. L., Viazzi, F., Venturelli, C., Brunori, G. and Garibotto, G. (2017). Toll-like receptor 4 signalling mediates inflammation in skeletal muscle of patients with chronic kidney disease. J Cachexia Sarcopenia Muscle 8(1): 131-144.
- Zhang, G., Liu, Z., Ding, H., Miao, H., Garcia, J. M. and Li, Y. P. (2017). Toll-like receptor 4 mediates Lewis lung carcinoma-induced muscle wasting via coordinate activation of protein degradation pathways. Sci Rep 7(1): 2273.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Verzola, D., Milanesi, S., Costigliolo, F. and Garibotto, G. (2019). Immunohistochemical Staining of TLR4 in Human Skeletal Muscle Samples. Bio-protocol 9(2): e3144. DOI: 10.21769/BioProtoc.3144.
Category
Immunology > Immune cell imaging > Confocal microscopy
Cell Biology > Tissue analysis > Tissue staining
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










