- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of BMEC Lines and in vitro BMEC-HSPC Co-culture Assays
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3079 Views: 7781
Reviewed by: Jia LiLokesh KalekarAbhijit Kale

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Murine Osteoblast and Osteoclast Co-culture on Demineralized Bone Paper for Bone Remodeling
Seema Amin [...] Jungwoo Lee
Jun 5, 2025 2225 Views

Assessing Human Treg Suppression at Single-Cell Resolution Using Mass Cytometry
Jonas Nørskov Søndergaard [...] James B. Wing
Aug 20, 2025 2912 Views

Isolation and Co-culture of Paneth Cells and Intestinal Stem Cells
Ryosuke Isotani [...] Toshimasa Yamauchi
Sep 20, 2025 3461 Views
Abstract
Endothelial cells (ECs) sustain the self-renewal and regeneration of adult hematopoietic stem and progenitor cells (HSPCs) via deployment of EC-derived paracrine factors, termed as angiocrine factors. Generation of durable ex vivo vascular niche that maintains EC identity and preserves the angiocrine profile of organ of origin offers platforms for in vitro dissection of the mechanism by which angiocrine factors execute their instructive function for stem cell maintenance and tissue regeneration. This protocol describes detailed methods to isolate primary bone marrow ECs (BMECs), to subsequently transduce lentiviral vector carrying myristoylated-Akt1 into primary BMECs, and to use the Akt1-BMECs to expand engraftable murine HSPCs. The BMEC-HSPC co-culture system serves as bioreactor prototype to generate scalable populations of the blood and immune systems.
Keywords: AngiocrineBackground
Hematopoietic stem cells (HSCs) are multipotent adult stem cells that can self-renew to replenish themselves and differentiate into all the lineages of the blood and immune system. HSC transplantation offers the best therapeutic cure for diseases such as acute myeloid leukemia, and served as cellular platform to correct the mutation of genetic blood disease via gene targeting. There are several sources of hematopoietic stem cells, adult bone marrow-derived HSCs, cord blood-derived HSCs, and granulocyte-colony stimulating factor (GCSF)-mobilized HSCs. Compared with the bone marrow-derived HSCs, cord blood HSCs can tolerate more HLA mismatching, and have better anti-leukemia activities, and are more readily available. Unfortunately, the HSC transplantation is still a risky procedure to perform and transplant-related mortality is partially due to the leukemia relapse and/or the incidence of infections that took place during the recovery phase of HSC transplantation; all of which are attributable to the low stem cell numbers in the donor cord blood. Therefore, identifying cellular and molecular approaches that can help expand bona fide HSCs that maintain self-renewal activity is of pivotal translational significance.
Endothelial cells safeguard the self-renewal and regeneration of adult HSCs in the bone marrow via deploying endothelial-derived paracrine factors, termed as angiocrine factors, such as KitL, SDF-1, Jagged-1 and Jagged-2, etc. (Poulos et al., 2013; Mendelson and Frenette, 2014; Rafii et al., 2016, Asada et al., 2017). Rafii et al. have pioneered in the technology of isolating adult human bone marrow endothelial cells (BMECs) and performing co-culture experiments to expand HSPCs (Rafii et al., 1994). The short life span of human BMECs and human umbilical venous endothelial cells (HUVECs) were strategically (Zhang et al., 2004) overcome via overexpression of the adenoviral E4ORF1 gene. The resulting cells, termed as E4-HUVECs, maintain the angiocrine profiles of primary HUVECs and are able to support the self-renewal of long-term repopulating mouse HSPCs and human cord blood stem cells (Seandel et al., 2008; Butler et al., 2010). E4ORF1 executes such functions partially via activation of Akt1 signaling pathway. We have thus generated constitutively active Akt1 by adding a myristoylation sequence 5’ to the ORF sequence that helps target the Akt1 at the cell membrane to undergo phosphorylation. Transduction of myristoylated Akt1 into primary mouse BMECs maintains their EC identities and preserves the angiocrine profiles (Kobayashi et al., 2010). This approach is useful for in vitro assays to dissect the mechanism through which BMECs support the self-renewal and expansion of HSPCs (Poulos et al., 2013; Hadland et al., 2015; Poulos et al., 2015; Guo et al., 2017). With the concept of endothelial cell heterogeneity, Akt1-BMECs will prove to be a complementary approach for in vivo studies that highlight the instructive role of different vascular beds for the differentiation of subpopulation of the blood and immune system.
This protocol is divided into the following parts:
- FACSAria II sorting of primary BMECs.
- Dynabeads isolation of primary BMECs.
- Lentiviral transduction of primary BMECs.
- In vitro BMEC-HSPC coculture.
Materials and Reagents
- Falcon tube, 15 ml (Corning, catalog number: 352096)
- Falcon tubes, 50ml (Corning, catalog number: 352098)
- Falcon® 100 mm TC-treated Cell Culture Dish, 20/Pack, 200/Case, Sterile (Corning, catalog number: 353003)
- Low retention tube, 1.5 ml (Fisher Scientific, catalog number, FisherbrandTM, catalog number: 02-681-320)
- Kimwipes (KCWW, Kimberly-Clark, catalog number: 34120)
- Parafilm (Bermis, catalog number: PM996)
- Amicon Ultra-0.5 Centrifugal Filter Unit (Merck, catalog number: UFC503096)
- Sterile 40 μm nylon mesh (Corning, catalog number: 352340)
- 6-tube magnetic stand (Thermo Fisher Scientific, catalog number: AM10055)
- 24-well plate (Corning, catalog number: 353047)
- 12-well plate (Corning, catalog number: 353043)
- T75 flasks (Corning, catalog number: 353136)
- Bio-Spin® P-30 Gel Columns, Tris Buffer (Bio-Rad Laboratories, catalog number: 7326232)
- DynabeadsTM Sheep anti-rat IgG (Thermo Fisher Scientific, catalog number: 11035)
- DPBS, 1x without calcium and magnesium (Corning, catalog number: 21-031-CV)
- DMSO, dimethyl sulfoxide (Sigma-Aldrich, catalog number: D2650)
- Alexa FluorTM 647 NHS Ester (Succinimidyl Ester) (Thermo Fisher Scientific, catalog number: A20006)
- Isothesia (Isoflurane) solution (Henry Schein Animal Health, catalog number: 029405)
- Isoflurane chamber, EZ anesthesia
- Oxygen (Tech Air)
- Potassium chloride, KCl, BioXtra, ≥ 99.0% (Sigma-Aldrich, catalog number: P9333)
- Calcium chloride dihydrate, CaCl2•2H2O (Sigma-Aldrich, catalog number: C3306)
- Magnesium chloride, MgCl2, anhydrous, ≥ 98% (Sigma-Aldrich, catalog number: M8266)
- Sodium bicarbonate, NaHCO3 (Sigma-Aldrich, catalog number: S5761-500G)
- Bovine Serum Albumin, lyophilized powder, essentially IgG-free, low endotoxin, BioReagent, suitable for cell culture (Sigma-Aldrich, catalog number: A2058)
- HBSS buffer: Hank’s Balanced Salt Solution, 1x without Calcium, Magnesium and Phenol Red (Corning, catalog number: 21-022-CV)
- Lentiviral myristoylated-Akt1: Virus titer is measured by Lenti-X p24 Rapid Titer Kit (Takara Bio, Clontech, catalog number: 632200)
- Fibronectin (Sigma-Aldrich, catalog number: F1141-5MG) (working concentration is 1 μg/ml in PBS)
- Trypsin (Corning, catalog number: 25-052-CI)
- Collagenase (Roche Diagnostics, catalog number: 11088793001)
- Dispase (Roche Diagnostics, catalog number: 04942078001)
- Polybrene (Sigma-Aldrich, catalog number: H9268-5G)
- Direct lineage depletion kit (Miltenyi Biotec, catalog number: 130-110-470)
- Accutase cell detachment solution (Corning, catalog number: 25-058-CI)
- StemSpan SFEM (Stem Cell Technologies, catalog number: 09650)
- Knockout serum replacement (Thermo Fisher Scientific, catalog number: 10828028)
- Recombinant human SCF, or KitL (PeproTech, catalog number: 250-03)
- UltraPureTM 0.5 M EDTA, pH 8.0 (Thermo Fisher Scientific, catalog number: 15575020)
- DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) (Thermo Fisher Scientific, catalog number: D1306)
- F-12 medium (Corning, Cellgro, catalog number: 10-080-CV)
- DMEM low-glucose medium (Corning, Cellgro, catalog number: 10-014-CV)
- Heat-inactivated FBS (Denville Scientific, catalog number: FB5001)
- Non-essential amino acid (Corning, Cellgro, catalog number: 25-025-CI)
- Penicillin/streptomycin/amphotericin (Corning, Cellgro, catalog number: 30-004-CI)
- 1 M HEPES (Corning, Cellgro, catalog number: 25-060-CI)
- Endothelial cell growth supplement (Alfa Aesar, catalog number: J64516)
- Heparin sodium 10 mg/ml (Sigma-Aldrich, catalog number: H3149-100KU)
- GlutaMAX (Thermo Fisher Scientific, catalog number: 35050061)
- Antibodies (Table 1)
- 8x stock concentration of collagenase/dispase solution (see Recipes)
- MACS buffer (see Recipes)
- Polybrene stock solution (see Recipes)
- EC complete medium (see Recipes)
Table 1. List of antibodies used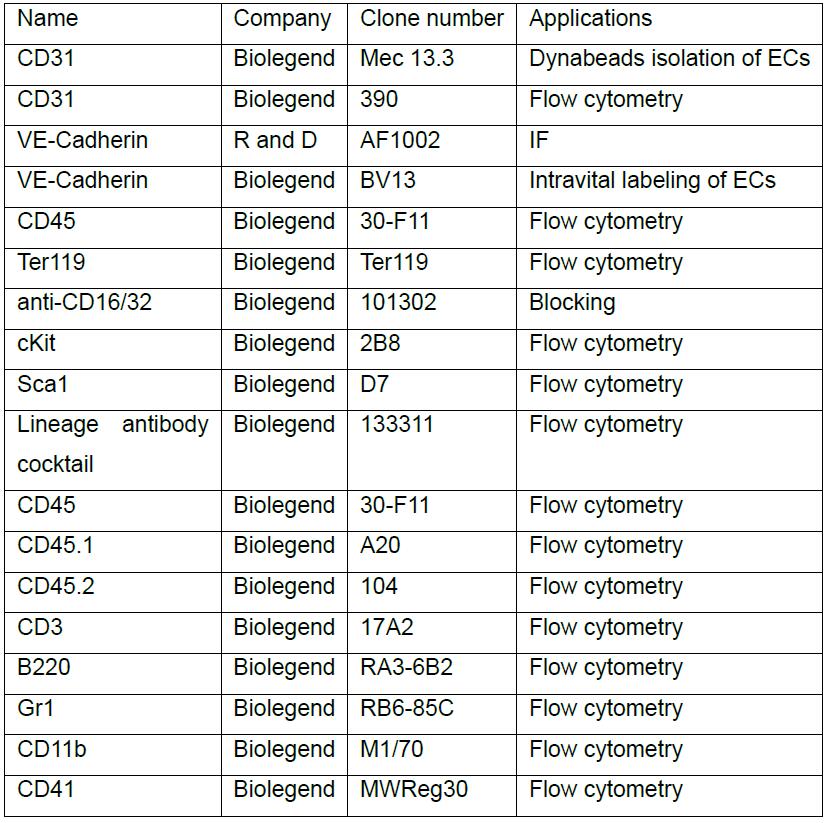
Equipment
- Pipettes, Denville Ultra EZpetteTM Pipette Starter Kit (Gray/Blue) (Denville Scientific, catalog number: P3960-SK)
- Scissors, Straight; Sharp-Blunt; 4.5" Length (Roboz Surgical Instrument, catalog number: RS-6800)
- Forceps, Student Grade Thumb Dressing Forceps 4.5" Serrated (Roboz Surgical Instrument, catalog number: 65-8100)
- Mortar and pestles. (VWR, catalog numbers: 89038-148 and 89038-164)
- 37 °C Orbital Shaker (Thermo Fisher Scientific, model: MaxQTM 4000)
- Centrifuge, SorvallTM LegendTM XT/XF Centrifuge Series (Thermo Fisher Scientific, model: SorvallTM LegendTM XT, catalog number: 75004505)
- FACS Aria II cell sorter (BD, model: FACSAria II)
- Laminar flow hood (The Baker Company, SterileGARD biological safety cabinets)
- NanoDrop Spectrometer (Thermo Fisher Scientific, model: NanoDropTM 1000, catalog number: ND-1000)
Procedure
- Prepare BV13-AF647 conjugated antibodies and measure the degree of labeling (DOL)
- Aliquot the AF647 dye into individual aliquots of 40 μg, by first resuspend 1 mg AF647 dye in 500 μl of DMSO, followed by aliquoting 20 μl into each tube and rotating desiccation.
- Concentrate antibodies to 1 mg/ml (100 μg total) in a 0.5 ml Amicon Ultra Filter Unit.
- Collect antibody by inverting the column into a fresh tube and spinning at 1,000 x g for 3 min.
- Bring the volume up to 100 μl with PBS.
- Add 10 μl of 1 M NaHCO3.
- Resuspend pellet of dye with antibody solution, incubate at 37 °C for 2 h.
- Pipette to mix every 20 min to evenly conjugate.
- When there is 5 min left to the conjugation, prepare the Bio-Spin® 30 (Bio-Rad) columns for purification.
- Swing tube to get slurry out the lid.
- Remove orange cap.
- Twist off bottom.
- Place in a 5 ml polystyrene Falcon tube to collect flow-through.
- Spin at 1,100 x g for 3 min in a swing bucket rotor.
- Load antibody/dye solution drop by drop onto the center of the resin.
- Place the column into a collection tube and spin for 1,100 x g for 5 min.
- Place into an amber tube and store at 4 °C.
- Calculate:
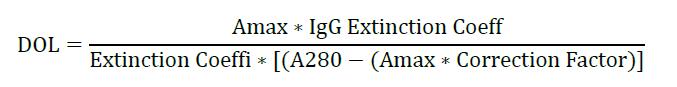
Note that for Amax and A280 are measured using a NanoDrop under the section “proteins and labels”. AF647 dye, the Amax is measured at 651 nm. Extinction Coeffi is 239,000, Correction Factor is 0.03, and Extinction Coefficient of typical IgG is 203,000.
For AF647,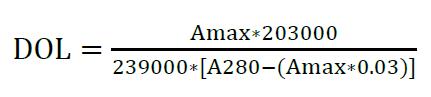
- Isolation of primary BMECs via cell sorting
- Prepare BV13-AF647 conjugated antibodies, at 1 mg/ml. Measure the degree of labeling (DOL). A good DOL to work with is between 4 and 8.
- Per mouse, prepare BV13-AF647 antibody by adding 25 μl of conjugated antibody in 75 μl of sterile PBS.
- Anesthetize the mouse using isofluorane and oxygen flow. Retroobitally injects the 100 μl BV13-AF647 antibody (25 μg) prepared at Step B2 into each mouse (Video 1). Video 1. Retroobital injection of BV13-AF647. After putting the mouse in the isoflurane chamber for 3 min, the mice become anesthetized (not shown in video). After confirming the state of anesthesia by pinching the in-between-toe area and lack of movement of mouse, the mouse was taken out from the isoflurane chamber and subjected to retro-orbital injection. Position the head of mouse closer to the needle. Firmly hold the eye area by placing the thumb and index fingers at each side of the eye ball. Using a downward motion, expose the eye ball as much as possible and meanwhile maintaining the eyeball in still position (This required placement of thumb and index fingers immediately adjacent of eye ball, but not further down below). Insert the needle from the anterior end of the eye to the area subneath the eyeball, (optional: one can inject the needle fully until the needle feels the bones behind the retroorbital plexus and retrieve the needle a little bit). Inject the 25 μg of BV13-AF647 diluted in 75 μl PBS (in total 100 μl volume) into the mouse. Quickly detach the needle and gently cover the eyes with eye lids to prevent bleeding. Mice were monitored for any signs of bleeding or other discomfort, which usually do not take place. (Study approval. All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee, New York, NY. All experimental procedures followed the IACUC guidelines. This video was made at Weill Cornell Medical College according to the guidelines of the IACUC of Weill Cornell Medical College, New York, New York, USA under protocol # 2009-0061.)
- Ten minutes later, euthanize the mouse.
- Quickly open the mouse, dissect out the 2 femurs and 2 tibias.
- Clean out the muscles using Kimwipes and scissors (Video 2). Video 2. Dissecting femurs and tibias. Expose and peel off the skin of legs using scissors. When dissecting out the femurs from mice, make sure to cut as much as possible at the end where femur and hip joints are attached, to preserve the intactness of femurs. After the femurs and tibias are dissected out from mice, excessive muscles are removed from the leg of mice using scissors (not shown in video). The femur and tibias was then separated using a gentle twisting motion. To further clear off the muscles from femurs, Kimwipes are used to scratch off the muscles, any residual muscle that is not easily detached using Kimwipes can be cut off using scissors. To clear off muscles from tibias, hold the tibias bottom up, the toe facing upward, with the back of foot facing toward the operator. Gently cut using scissors at the wrist area to expose the skin, then the skin and muscles can all be peeled off by pulling the toe downward and simultaneously pushing the tibias bone upward (see video for detailed techniques). (Study approval. All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee, New York, NY. All experimental procedures followed the IACUC guidelines. This video was made at Weill Cornell Medical College according to the guidelines of the IACUC of Weill Cornell Medical College, New York, New York, USA under protocol # 2009-0061.)
- Prepare 1x collagenase/dispase solution by diluting the 8x collagenase/dispase stock with 1x HBSS buffer.
- Homogenize femurs and tibias using a mortar and pestle. Use the pestle to firmly grind the bone tissues in a circular motion, clockwise for 25 times, and counter clockwise for 25 times. Make sure all the marrow tissue is released.
- Add 5 ml of 1x collagenase/dispase/HBSS buffer to the mortar, and transfer all the supernatant and the bone tissues into a 15 ml Falcon tube.
- Seal the Falcon tube with parafilm and place the tube on an orbital shaker at 37 °C. Shake for 15 min.
- Add 10 ml MACS buffer to stop the enzymatic digestion.
- Filter cells through a sterile 40 μm nylon mesh (cell strainer) and centrifuge flow through at 500 x g for 5 min.
- Aspirate supernatant carefully.
- Carry out lineage depletion using direct lineage depletion kit (Miltenyi Biotech). Collect the Lin- cells from the flow through.
- Resuspend cell pellet in 50 μl of MACS buffer.
- Block with TruStain mouse block (FcR block, or anti-CD16/32 antibody) at 1:50 dilutions for 5 min on ice.
- Add 1 μl CD31 and 1 μl CD45 antibodies into the cells and stain on ice for 25 min.
- Add 10 ml MACS buffer to wash.
- Prepare resuspenstion solution (PBS + 2 mM EDTA + DAPI). After taking out the supernatant, add 0.4 ml (PBS + 2 mM EDTA + DAPI) into each tube to resuspend the pellet.
- Sort the BMECs gated as DAPI-CD45-CD31+VE-Cadherin+ cells at 85 μm nozzle using BD FACSAria II (Figure 1A). Alternatively, when the intravital labeling of the BV13-AF647 was not performed, BMECs can be sorted as DAPI-CD45-CD31+ cells (Figure 1B).
- From one 2-month old mouse, we can obtain about 20,000-30,000 BMECs.
- Seed cells into one well of 24-well plate, in 0.5 ml of mouse EC complete medium.
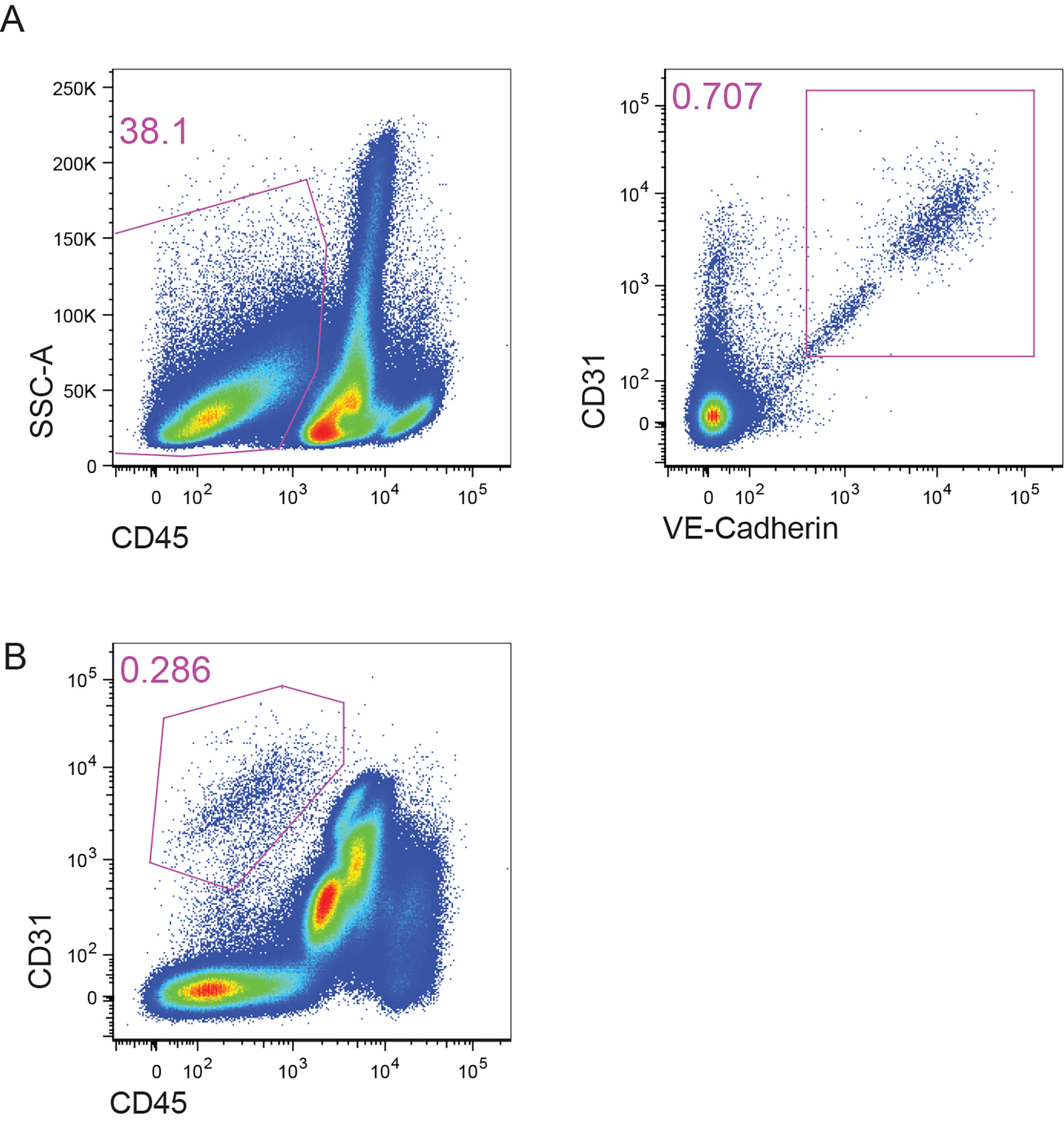
Figure 1. Gating of BMECs for sorting. A. The Lin- cells were further gated on CD45- and subsequently CD31+VE-Cadherin+ population. The obtained cell population are primary BMECs. B. Alternatively, BMECs can be sorted as CD31+CD45- cells.
- Isolation of primary BMECs via CD31-Dynabeads
- Day 1: Coat sheep anti-rat Dynabeads with rat anti-mouse CD31 (Clone 13.3) antibody
- Use 10 μl beads for femurs and tibias from one mouse.
- Wash beads three times each with 1 ml MACS buffer on magnetic rack.
- Resuspend beads in 200 μl MACS buffer.
- Add 4.8 μl of rat anti-mouse CD31 antibody.
- When working with several mice’s bone marrow samples, scale up the beads volume at Step B1a and the antibody volume at Step B1d, but keep the MACS buffer volume at 200 μl.
- Incubate beads and antibody with gentle mixing for 1 h at room temperature and then keep mixing at 4 °C overnight.
- Day 2: Isolate primary BMECs
- Euthanize the mouse.
- Quickly open the mouse, dissect out the 2 femurs and 2 tibias.
- Clean out the muscles using Kimwipes and scissors.
- Prepare 1x collagenase/dispase solution by diluting the 8x collagenase/dispase stock with 1x HBSS buffer.
- Homogenize femurs and tibias using a mortar and pestle. Use the pestle to firmly grind the bone tissues in a circular motion, clockwise for 25 times, and counter clockwise for 25 times. Make sure all the marrow tissue is released.
- Add 5 ml of 1x collagenase/dispase/HBSS buffer to the mortar, and transfer all the supernatant and the bone tissues into a 15 ml Falcon tube.
- Cap the Falcon tube, and then seal the Falcon tube with parafilm. Place the tube on an orbital shaker at 37 °C. Shake for 15 min.
- Add 10 ml MACS buffer to stop the digestion.
- Filter cells through a sterile 40 μm nylon mesh (cell strainer) and centrifuge flow through at 500 x g for 5 min.
- Aspirate supernatant carefully.
- Resuspend the cell pellet in 0.5 ml MACS buffer in a low retention tube.
- Wash beads 3 times in MACS buffer to remove excessive antibody. Resuspend beads in 50 μl MACS buffer per 10 μl beads volume at Step B1a of Day 1.
- Add CD31-coated beads and incubate for 45 min at 4 °C, with gentle shaking.
- Collect the bead-bound cells using a magnet and wash 5 times in MACS buffer.
- Precoat one well of 12-well plate using fibronectin at 1 μg/ml in PBS.
- Resuspend the beads using 1 ml mouse EC complete medium and transfer to a 12-well plate for culture. After Dynabeads-CD31 enrichment, we put all the CD31+ cells obtained from one mouse’s 2 femurs and 2 tibias into one well of 12-well plate pre-coated with fibronectin.
- It is recommended to use an appropriate amount of Dynabeads to enrich CD31+ cells. Excessive beads tend to accumulate at the bottom of the well plate and prevent the cells from attaching, decreasing the yield (Figure 2).
- The Dynabeads can be removed by enzymatic digestion using trypsin or accutase cell detachment medium. Alternatively, as cells divide or during cell passages, Dynabeads will gradually disappear.
- Comparison between the cell sorting techniques and the Dynabeads enrichment approach: Pure population of BMECs will be obtained from FACS sorting, albeit at low cell number. For generation of stable lines of Akt1-BMECs, Dynabeads approach yields more cells and better viability for seeding. Any impurities at the initial step can be taken care of at the later steps via FACS sorting.
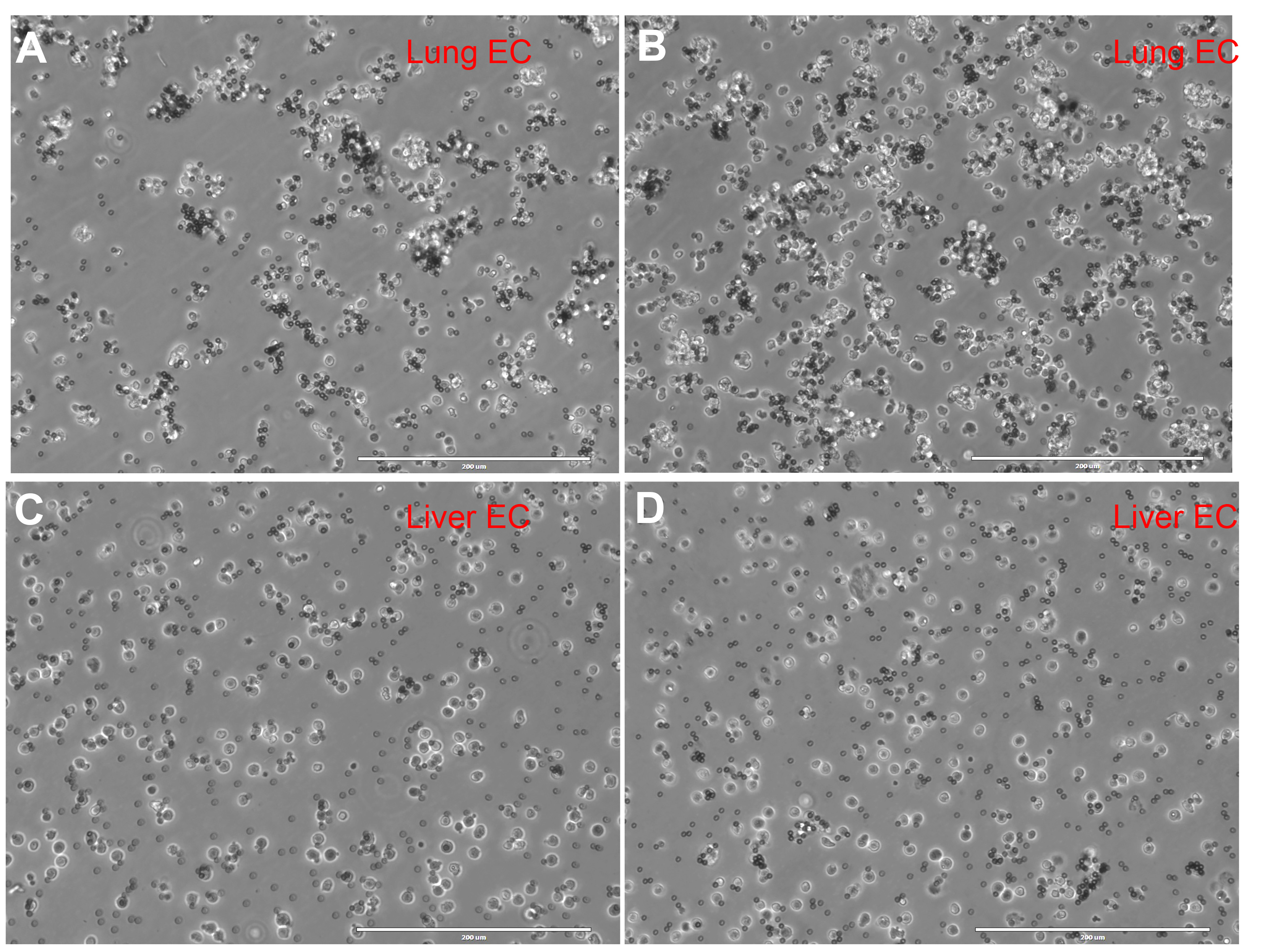
Figure 2. Representative images of primary mouse CD31+ cells post Dynabeads enrichment. A-B. Bright field image of Dynabeads enriched C31+ lung ECs. C-D. Bright field image of Dynabeads enriched C31+ liver ECs. There are few free Dynabeads floating in the cell culture dish, making it easier for cells to attach. Scale bars =200 μm.
- Day 1: Coat sheep anti-rat Dynabeads with rat anti-mouse CD31 (Clone 13.3) antibody
- Lentiviral transduction of primary BMECs and cell passages
- Day 1
- Precoat the plate with fibronectin/PBS solution (1:1,000 dilution of fibronectin in sterile PBS) for 30 min at room temperature in the laminar flow hood.
- Aspirate off the fibronectin coating.
- Seed the primary BMECs in mouse EC complete medium in the precoated wells. Transform the number of sorted events and divide by 3 to come up with the actual cell number, and seed within smaller well plates, such as 24-well plate. From one mouse’s 2 femurs and 2 tibias, we put the Dynabeads enriched CD31+ cells into one well of 12-well plate. We put the FACS purified BMECs into one well of 24-well plate.
- Day 2
- One day after plating, some of the cells should have attached. Cells tend to attach at the center and the very edge of the plate.
- Add polybrene solution into the medium to a final concentration of 4 μg/ml.
- Let sit for 5 min at 37 °C in the cell incubator.
- Add 2,500 pg of virus for one well of 24-well plate. Scale up or down as needed.
- Day 3
Add 0.5 ml of fresh medium into the transduced wells (24-well plate). - Days 4-6
- Small colonies should be growing out, at the center and at the very edge of the plate.
- Gently change the medium after emergence of such colonies.
- Days 7-20
- Change the medium every 3 days, until it reaches more than 80% confluence.
- Around Day 20, passage the cells into a bigger well place, at 1:2. This first-time passage is tricky and should be done very carefully. It is normal to lose cells after replating them into a new well. Trypsin is better than accutase. The primary mouse ECs have been sitting in the same well for 3-4 weeks, and are very firmly attached to the bottom of the plate. Using accutase takes at least 30 min to detach the cells, and thus very harmful. 0.05% trypsin detaches the cells faster and preserves the cells significantly better than accutase. Add 1ml of 0.05% trypsin into 1 well of a 12-well plate, and incubate at 37 °C for 7 min, or until the cells round up when observed under a microscope. Gently detach the cells using pipettes.
- Day 20 and beyond
- Change the medium every 3-4 days until the cells reach more than 80% confluence.
- Passage 1:2 into one well of a 6-well plate.
- Repeat Steps C6a-C6b, until the cells reach confluence in T75 flasks. Confirm the purity of cultured Akt1-mouse ECs and purity by sorting if necessary.
- Cryopreserve aliquots of early passage Akt1-mouse ECs for future usage.
- Timeline: After CD31-Dynabeads isolation of primary ECs, the cells were seeded into 12-well plate. After lentiviral transduction and subsequent culture, it takes about 3-4 weeks to reach one confluent well in a 12-well plate. The subsequent cultures and passages are easier than the first cell passage. It usually takes 2 months to reach confluence in one T75 flask of cells.
- Use trypsin, not accutase for cell passaging of murine ECs, especially the first time cell passage.
- Dilute 1:2 when doing the initial rounds of cell passage.
- Due to intrinsic endothelial cell heterogeneity, it is recommended to generate multiple lines of ECs and compare their functions.
- Especially for the earlier passages of ECs, there are multiple colonies in the same well, and the cells look heterogenous (Figure 3A). After passaging for more than 10 times, the cells tend to become more and more homogenous (Figure 3B).
- Most of the times, I used 20% oxygen culture conditions when generating the murine ECs lines. 5% oxygen condition should be explored for better yield.
- Using the above protocols, mouse EC lines from lung, liver, and brain etc. have been successfully generated.
- Key events: If you see cells attach after 2-3 days post seeding, and especially observe a few colonies growing out and persistently gets larger after lentiviral transduction around Days 6-7 post seeding, most likely the cells are going to grow fine. A small number of colonies go a long way, and they proliferate remarkably to generate a murine EC line with scalable cell numbers.
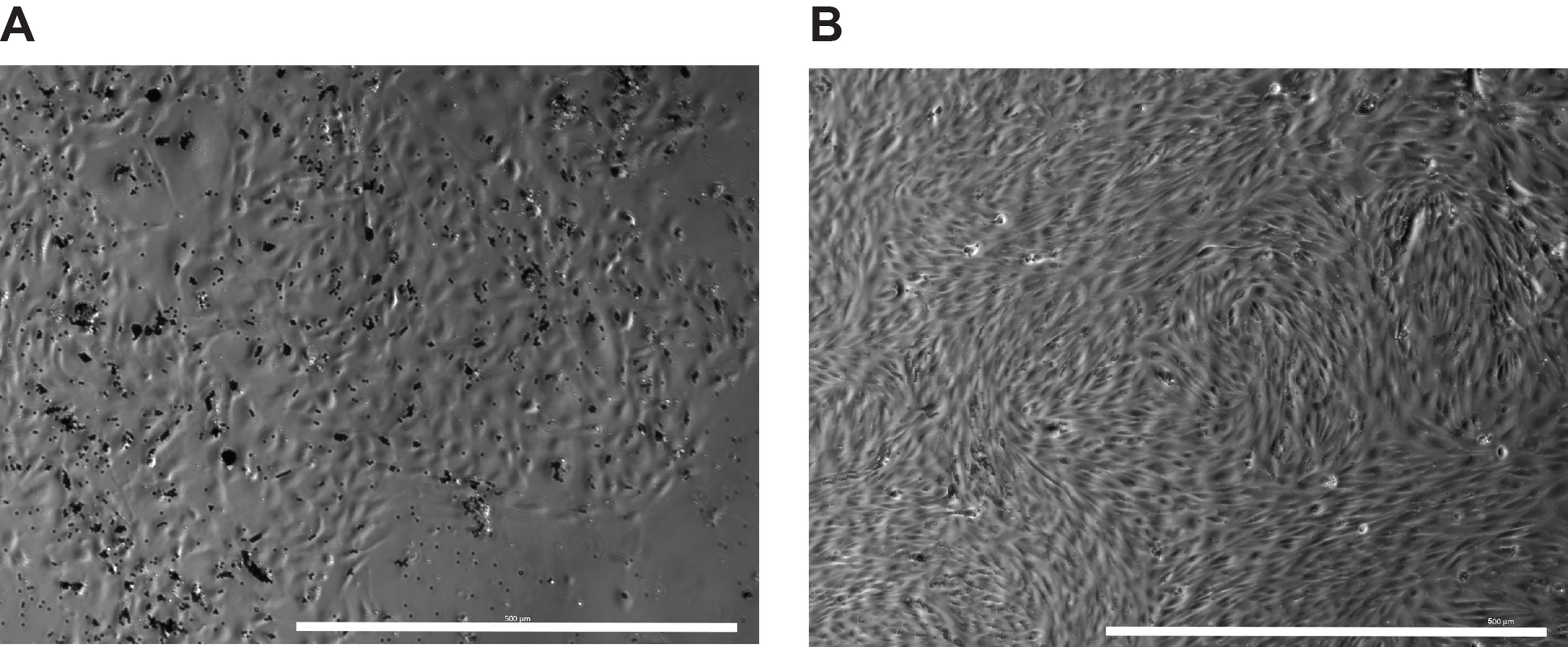
Figure 3. Representative images of Akt1-BMECs at different stages. A. At day 10 post Akt1 transduction of the Dynabeads enriched CD31+ primary BMECs. Note that several colonies of ECs exist in the well (Scale bar = 500 μm). B. After cell passaging for more than 10 times, the resulting Akt1-BMECs become homogenous (Scale bar = 500 μm).
- Day 1
- In vitro Akt1-BMEC and HSPC coculture assays
- Seed BMECs into a 12-well plate and let it grow into confluence in mouse EC complete medium.
- Euthanize Mice. Dissect femurs and tibias out.
- Add 5 ml of MACS buffer into the mortar and then homogenize femurs and tibias (Video 3).
- Filter the supernatant through a 40 μm cell strainer into a 50 ml Falcon tube (Video 3).Video 3. Homogenizing femurs and tibias to obtain bone marrow cells. Put the 2 femurs and 2 tibias dissected from one mouse into the mortar, and add 5 ml of cold, sterile MACS buffer. Firmly hold the pestle and apply downward force to grind up the femurs and tibias. After the femurs and tibias are broken into pieces, use circular motion to fully release the bone marrow cells within the bone cavities. Apply circular grinding motion for 25 times clockwise and 25 times counter clockwise. For the isolation of BMECs as we discussed in this protocol, both the white bony tissues and the cell suspension are collected into the 15 ml Falcon tube, followed by enzymatic digestion. For collecting hematopoietic cells to retrieve Lin- cells for coculture, collect 5 ml of supernatant containing the cell suspension and immediately filter through a 40 μm cell strainer into a 50 ml Falcon tube. Add 5 ml MACS buffer into the white bony remains and apply circular grinding motion. Collect the supernatant until the red bone marrows have been completely rinsed out, leaving only white bone tissues. Usually, it takes 3 rounds of 5 ml MACS buffer and circular grinding to complete the homogenization processes. No enzymatic digestion (collagenase and dispase) is needed. In total, for collecting the hematopoietic stem cells, we have about 15 ml cell suspension in MACS buffer, ready for downstream processing.
- Repeat Steps D3-D4 two more times until all the pieces of bone marrow appear white.
- Enrich lineage-negative cells (Lin-) cells using direct lineage depletion kit.
- To prepare the Akt1-BMECs for co-culture, first aspirate off the mouse EC complete medium from the wells.
- Wash the Akt1-BMECs once with 1x PBS without calcium/magnesium.
- Prepare the mouse HSPC culture medium using StemSpan supplemented with 20 ng/ml sKitL, knockout serum replacement, penicillin/streptomycin/amphotericin and glutaMAX.
- Resuspend the Lin- cells in 0.1 million/ml StemSpan medium. For example, if 3 wells of 12-well plate of coculture are needed, resuspend 0.3 million of Lin- cells in 3 ml of StemSpan medium.
- Day 0: Add 0.1 million of Lin- cells into one well of a 12-well plate. This is considered as Day 0.
- Day 2: Add 1 ml of StemSpan medium supplemented with sKitL into each well.
- Prepare a second 12-well plate of Akt1-BMECs and let it become confluence by Day 4.
- Day 4: Gently collect the floating hematopoietic cells and spin them down at 500 x g for 5 min. Resuspend into 1ml of StemSpan medium and distribute into 1 well of 12-well plate as on Day 0. Add 1 ml each of fresh medium into the old wells of BMECs with Lin- cell coculture. Keep both the old wells and the new wells for culture.
- Day 6: Add 1 ml of fresh medium to both the new well and old wells.
- Day 7 and onward: Cell collection, data analysis and downstream functional assays.
- On Day 7, collect the floating hematopoietic cells. Add 0.3 ml of accutase into each well of the 12-well plate to detach both the attached HSPCs and the BMECs.
- For calculation of total expanded HSPCS: the total hematopoietic cell number are counted.
- Enrich Lin- cells using direct lineage depletion kit and count the resulting Lin- cell numbers.
- And then stain the Lin- cells with CD45, c-Kit, and Sca1 antibodies to obtain the frequency of cKit+Sca1+Lin- HSPCs among the Lin- cells.
- Calculate the total numbers of expanded HSPCs using the Lin- cell number and the percentage of HSPC among Lin- cells.
- Phenotyping the lineage cells after 7 days of co-culture
- Take out approximately 1 million total expanded cells and wash with PBS once. After decanting the supernatant, resuspend the cells in 50 μl of MACS buffer. Block the cells with mouse FcR block at 1:50 dilution for 10 min on ice. Add lineage antibodies (1 μl of each undiluted antibody) including CD45, Gr1, CD11b, B220, CD3, CD41 into the cell suspension. Stain the antibodies and the cells on ice for 25 min.
- Wash off the excessive and unspecific bound antibodies using 1 ml of MACS buffer. Finally, resuspend the cells in 0.4 ml (PBS + 2 mM EDTA + DAPI) solution for flow cytometry analysis.
- If the cells are not analyzed on the same day when the staining is done, after washing off the excessive antibodies, fix the cells in 200 μl of 1% PFA/PBS solution at room temperature for 3 min. Then wash the PFA using 1 ml of MACS buffer. Finally, resuspend the cells in 0.4 ml of (PBS + 2 mM) buffer. The stained cells can be stored for 2 days before flow cytometry analysis.
- The myeloid cells are defined as CD45+Gr1+ or CD45+CD11b+, T cells are defined as CD45+CD3+ cells, and B cells are defined as CD45+B220+ cells. Megakaryocyte lineages are defined as CD45+CD41+ cells.
- Functional analysis of the HSPCs can be performed using in vitro methylcellulose assays and competitive transplantation assays. For the methylcellulose assays, sort out 350 cKit+Sca1+Lin- cells (events) in 300 μl of StemSpan medium. Then add the cells into 3 ml of thawed methylcult, and split into 2 low-attachment Petri dish. On Day 8 post culture, analyze the resulting colony number and colony type.
- For competitive transplantation assays, irradiate CD45.1 mice on Day 6 of coculture at 9 Gy. On Day 7, count 0.5 million of CD45.2 co-cultured hematopoietic cells and mix with 0.5 million of CD45.1 bone marrow mononucleated cells (BMMNCs), and next retroorbitally inject into the CD45.1 mice. The BMECs are not sorted out when performing such competitive transplantation assays, and will not engraft into the CD45.1 mice. At 4, 8, 12 and 16 weeks post-transplantation, analyze the peripheral chimerism and lineage differentiation potential for CD45.2 cells.
Data analysis
Please refer to the methods sections named “In vitro BMEC-HSPC coculture assays’ in the article (Guo et al., 2017) and Figures 2A-2K for the data analysis.
Recipes
- 8x stock concentration of collagenase/dispase solution
- Prepare buffer to resuspend the enzymes in: 500 ml PBS, with 5 mM KCl, 10 mM HEPES, 2 mM CaCl2, 1.3 mM MgCl2. Filter to sterilize
- Take a sterile bottle, add 2.5 g collagenase A, and 1 g Dispase. Add 125 ml of buffer prepared in step a; Mix to resuspend. Do not filter. The solution is too sticky to be filtered
- MACS buffer
PBS
2 mM EDTA
0.1% BSA
Penicillin/streptomycin/amphotericin (Final concentration: 1x. The stock is 100x, dilute 1:100 when making MACS buffer)
Filter to sterilize - Polybrene stock solution
Prepare stock concentration using ddH2O at 4 mg/ml
Filter to sterilize in a laminar flow hood.
Final working concentration: 4 μg/ml-8 μg/ml depending on the cell types - Mouse EC complete medium
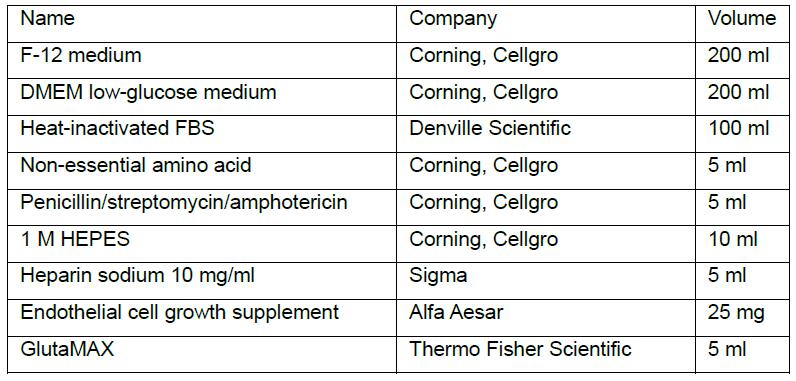
Note: Prepare heparin sodium powder to a stock concentration of 10 mg/ml using F-12 medium. Filter to sterilize.
Acknowledgments
SR is supported by the Ansary Stem Cell Institute, the Starr Foundation Tri-Institutional Stem Cell core project, the Tri-Institutional Stem Cell Initiative (TRI-SCI 2013-032, 2014-023, 2016-013), the Empire State Stem Cell Board, and New York State Department of Health grants, and by grants from the NIH R01 (DK095039, HL119872, HL128158, HL115128, HL099997) and U54 CA163167.
Competing interests
The authors declare no conflict of interest.
Ethics
All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee, New York, NY. All experimental procedures followed the IACUC guidelines. The videos were made at Weill Cornell Medical College according to the guidelines of the IACUC of Weill Cornell Medical College, New York, New York, USA under protocol # 2009-0061).
References
- Asada, N., Takeishi, S. and Frenette, P. S. (2017). Complexity of bone marrow hematopoietic stem cell niche. Int J Hematol 106(1): 45-54.
- Butler, J. M., Nolan, D. J., Vertes, E. L., Varnum-Finney, B., Kobayashi, H., Hooper, A. T., Seandel, M., Shido, K., White, I. A., Kobayashi, M., Witte, L., May, C., Shawber, C., Kimura, Y., Kitajewski, J., Rosenwaks, Z., Bernstein, I. D. and Rafii, S. (2010). Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 6(3): 251-264.
- Guo, P., Poulos, M. G., Palikuqi, B., Badwe, C. R., Lis, R., Kunar, B., Ding, B. S., Rabbany, S. Y., Shido, K., Butler, J. M. and Rafii, S. (2017). Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J Clin Invest 127(12): 4242-4256.
- Hadland, B. K., Varnum-Finney, B., Poulos, M. G., Moon, R. T., Butler, J. M., Rafii, S. and Bernstein, I. D. (2015). Endothelium and NOTCH specify and amplify aorta-gonad-mesonephros-derived hematopoietic stem cells. J Clin Invest 125(5): 2032-2045.
- Kobayashi, H., Butler, J. M., O'Donnell, R., Kobayashi, M., Ding, B. S., Bonner, B., Chiu, V. K., Nolan, D. J., Shido, K., Benjamin, L. and Rafii, S. (2010). Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol 12(11): 1046-1056.
- Mendelson, A. and Frenette, P. S. (2014). Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med 20(8): 833-846.
- Poulos, M. G., Guo, P., Kofler, N. M., Pinho, S., Gutkin, M. C., Tikhonova, A., Aifantis, I., Frenette, P. S., Kitajewski, J., Rafii, S. and Butler, J. M. (2013). Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep 4(5): 1022-1034.
- Poulos, M. G., Crowley, M. J. P., Gutkin, M. C., Ramalingam, P., Schachterle, W., Thomas, J. L., Elemento, O. and Butler, J. M. (2015). Vascular platform to define hematopoietic stem cell factors and enhance regenerative hematopoiesis. Stem Cell Rep 5(5): 881-894.
- Rafii, S., Butler, J. M. and Ding, B. S. (2016). Angiocrine functions of organ-specific endothelial cells. Nature 529(7586): 316-325.
- Rafii, S., Shapiro, F., Rimarachin, J., Nachman, R. L., Ferris, B., Weksler, B., Moore, M. A. and Asch, A. S. (1994). Isolation and characterization of human bone marrow microvascular endothelial cells: hematopoietic progenitor cell adhesion. Blood 84(1): 10-19.
- Seandel, M., Butler, J. M., Kobayashi, H., Hooper, A. T., White, I. A., Zhang, F., Vertes, E. L., Kobayashi, M., Zhang, Y., Shmelkov, S. V., Hackett, N. R., Rabbany, S., Boyer, J. L. and Rafii, S. (2008). Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc Natl Acad Sci U S A 105(49): 19288-19293.
- Zhang, F., Cheng, J., Hackett, N. R., Lam, G., Shido, K., Pergolizzi, R., Jin, D. K., Crystal, R. G. and Rafii, S. (2004). Adenovirus E4 gene promotes selective endothelial cell survival and angiogenesis via activation of the vascular endothelial-cadherin/Akt signaling pathway. J Biol Chem 279(12): 11760-11766.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Guo, P. and Rafii, S. (2018). Generation of BMEC Lines and in vitro BMEC-HSPC Co-culture Assays. Bio-protocol 8(21): e3079. DOI: 10.21769/BioProtoc.3079.
Category
Stem Cell > Adult stem cell > Hematopoietic stem cell
Developmental Biology > Cell growth and fate > Proliferation
Cell Biology > Cell isolation and culture > Co-culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









