- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Staining the Germline in Live Caenorhabditis elegans: Overcoming Challenges by Applying a Fluorescent-dye Feeding Strategy
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3077 Views: 7285
Reviewed by: Neelanjan BoseJuan Facundo Rodriguez AyalaSwati Jalgaonkar

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Blinded Visual Scoring of Images Using the Freely-available Software Blinder
Steven D. Cothren [...] Jessica H. Hartman
Dec 5, 2018 6428 Views

Use of Optogenetic Amyloid-β to Monitor Protein Aggregation in Drosophila melanogaster, Danio rerio and Caenorhabditis elegans
Prameet Kaur [...] Nicholas S. Tolwinski
Dec 5, 2020 5765 Views

Visualization and Purification of Caenorhabditis elegans Germ Granule Proteins Using Proximity Labeling
Hannah L. Hertz [...] Wen Tang
Apr 20, 2022 4214 Views
Abstract
C. elegans provides a tractable model organism for studying germline cell biology. Microscopy experiments are relatively facile, as this worm is transparent and germline development can be observed in real-time using DIC microscopy and/or fluorescent transgenes. Despite these many tools, robust staining techniques for imaging germ cells in live worms have been more elusive, due to the tough outer cuticle of the worm, which impairs staining efficiency. This limitation has restricted the spectrum of probes that can be used to investigate reproductive cell biology in C. elegans. Building on previous approaches, I recently applied a fluorescent-dye feeding strategy to reproducibly label organelles and monitor physiological changes in germlines of living C. elegans. In this approach, fluorescent dyes are initially introduced into the agar plates and bacterial lawns on which worms are subsequently cultured. After worms are grown on the dyed plates, oocytes show staining patterns consistent with verified transgenic markers. Thus, this approach offers an effective solution for labeling difficult-to-stain tissues in live worms, and establishes an entry point for incorporating new probes and sensors into analyses of C. elegans germline biology.
Keywords: C. elegansBackground
Animal reproduction is one of the most fascinating topics in biology. Understanding how germ (reproductive) cells are able to give rise to offspring with each new generation has intrigued scientists, as well as the general public, for centuries. With recent advances in imaging techniques and tools, researchers have been able to observe oocytes (female germ cells) and sperm (male germ cells) in extraordinary detail, revealing fundamental mechanisms that allow germ cells to execute their specialized functions. Model organism research, in particular, has afforded the opportunity to combine live-animal imaging with genetic analyses to tease apart relevant biological pathways important for germ cell regulation. The nematode C. elegans, which exists as a hermaphrodite that continually self-fertilizes throughout young-adulthood (Klass, 1977), presents a powerful experimental system for studying important questions in reproductive biology. Recently, I utilized the C. elegans germline to explore how signs of aging are reversed from one generation to the next (Bohnert and Kenyon, 2017). These studies required the use of dyes to track various aspects of oocyte physiology, including lysosomal acidity and mitochondrial membrane potential. Unfortunately, dyes incubated with C. elegans in liquid culture were inefficient at penetrating the worm’s outer cuticle, which is highly-impervious (Page and Johnstone, 2007) and precludes robust staining. To circumvent this obstacle, I applied a dye-feeding approach (Hermann et al., 2005; Schaheen et al., 2006) in an attempt to stain and image intact, full germlines of live C. elegans. When worms were grown on agar plates and food (E. coli OP50) to which dyes had previously been added, the germline, in addition to other adult tissues, could be clearly and accurately stained (Bohnert and Kenyon, 2017). Here, I describe this feeding-based strategy for staining the germline of live C. elegans, which can in principle be applied to other dyes that have difficulty penetrating the worm cuticle.
Materials and Reagents
- Graduated cylinders (Nalgene; Fisher, catalog number: 02-540-270)
- Plastic beakers (Nalgene; Fisher, catalog number: 02-591-10H)
- 15 ml Falcon tubes (Fisher, catalog number: 14-959-70C)
- 15 ml bacterial culture test tubes (Fisher, catalog number: 14-956-9C)
- 1.5 ml microcentrifuge tubes (Fisher, catalog number: 05-408-129)
- Weighing boats (Fisher, catalog number: 08-732-112)
- Pipet tips (Fisher, catalog numbers: 02-707-401, 02-707-415, and 02-707-436)
- Serological pipets (Fisher, catalog number: 13-676-10J and 13-676-10K)
- 35-mm Petri dishes (Lab Sciences, catalog number: 9333-35NV)
- 100-mm Petri dishes (Fisher, catalog number: FB0875712)
- Rubbermaid plastic container (Amazon, catalog number: B00YQAGNGK)
- Foil (any household variety works)
- Empty 1 mm gel cassette (Thermo Fisher, catalog number: NC2010)
- Razor blade (Fisher, catalog number: 18-100-970)
- Microscope slides (Fisher, catalog number: 12-518-100B)
- Microscope slide coverslips (Fisher, catalog number: 12-541B)
- C. elegans strains; for wild-type, use Bristol N2 (Caenorhabditis Genetics Center)
- E. coli OP50 strain (Caenorhabditis Genetics Center)
- Sodium chloride, NaCl (Fisher, catalog number: S271-10)
- Peptone (Fisher, catalog number: BP1420-500)
- Agar (Fisher, catalog number: BP1423-500)
- Calcium chloride dihydrate, CaCl2•2H2O (Fisher, catalog number: C79-500)
- Cholesterol (Alfa Aesar, catalog number: A11470-18)
- Ethanol (Fisher, catalog number: BP2818-500)
- Magnesium sulfate, MgSO4 (Alfa Aesar, catalog number: 33337-36)
- Potassium phosphate monobasic, KH2PO4 (Fisher, catalog number: P285-500)
- Potassium phosphate dibasic, K2HPO4 (Fisher, catalog number: P288-500)
- Sodium hydroxide, NaOH (Fisher, catalog number: S318-1)
- Household bleach (Chlorox)
- LB Broth, Miller (Fisher, catalog number: BP1426-2)
- Agarose (Fisher, catalog number: BP1356-100)
- Levamisole hydrochloride (Acros Organics, catalog number: 187870100)
- Carbenicillin (Fisher, catalog number: BP2648-1)
- IPTG (Fisher, catalog number: BP1755-10)
- Dyes
Note: The following dyes have been tested and verified to work with this protocol, but others may be suitable as well.- LysoTracker Red DND-99 (Life Technologies, catalog number: L7528)
- MitoTracker Deep Red FM (Life Technologies, catalog number: M22426)
- DiOC6(3) (Life Technologies, catalog number: D273)
- 1 M CaCl2 (see Recipes)
- 5 mg/ml cholesterol (see Recipes)
- 1 M MgSO4 (see Recipes)
- 1 M KPO4 pH 6.0 (see Recipes)
- 100 mg/ml carbenicillin (see Recipes)
- 1 M IPTG (see Recipes)
- NG agar (see Recipes)
- Dye stock solutions (see Recipes)
- LB medium (see Recipes)
- Worm bleaching solution (see Recipes)
- Agarose pads (see Recipes)
- Levamisole solution (see Recipes)
Equipment
Note: The following types of equipment will be needed for this protocol. Though the exact models are not absolutely necessary, I have included information on the specific units that my lab and I have used.
- Glass flasks (Kimble; Fisher, catalog number: 02-543-21)
- Glass beakers (Kimble; Fisher, catalog number: 02-555-2)
- Glass bottles (Pyrex; Fisher, catalog numbers: 06-414-1B, 06-414-1C, and 06-414-1D)
- Stir bar (Fisher, catalog number: 14-513-82)
- Pipets (Gilson; Fisher, catalog number: F167900G)
- Motorized pipet controller (Fisher, catalog number: FB14955202)
- Microwave (any household variety works)
- Autoclave (Steris, model: AMSCO 3000 series)
- Hotplate stirrer (Thermo Fisher, catalog number: 88880004)
- Shaker (Amerex Instruments, GYROMAX, model: 737R)
- Tabletop centrifuge (Eppendorf, model: 5804R)
- Vortex (Thermo Fisher, catalog number: 88880017TS)
- Hood
- Tube revolver (Thermo Fisher, catalog number: 88881001)
- Bacteria incubator (Fisher, catalog number: 15-103-0518)
- Worm incubator (Thermo Fisher, catalog number: PR205745R)
- Confocal microscope
We have used various microscope systems to image stained worms. In principle, any confocal system with appropriate filter sets for the selected dye(s) is suitable. I recommend using a 40X objective to image individual germlines. Examples of microscope systems that my lab and I have used include:- Leica, model: TCS SP8 with white light laser
- Nikon, model: CSU-Series
Software
- Imaging software
This will depend on the microscope system that is used. For example, the Leica TCS SP8 is equipped with LAS X software (https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/), whereas Nikon confocal microscopes have NIS Elements software (https://www.nikoninstruments.com/Products/Software). As with the microscope system, many different types of imaging software are in principle compatible with this protocol. - ImageJ
ImageJ can be used for image analysis (NIH; https://imagej.nih.gov/ij/).
Procedure
- Prepare 25 ml of NG agar in a flask per a standard recipe (see below) (Stiernagle, 2006). Include a stir bar.
- Autoclave NG agar using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min.
- Cool the NG agar. Add the post-autoclave ingredients from sterile stocks (see Recipes below) and stir on a hot-plate (Figure 1A).
- Once all ingredients have been mixed, add the dye. Tested dyes have been verified to work at a final concentration of 2 μM. Different concentrations may work for other dyes; when trying new dyes, it is a good idea to compare test plates of various dye concentrations for staining efficiency. Using less dye allows more plates to be made from the same starting amount of dye, which is cost-effective. However, a lower dye concentration also results in weaker staining, so the trade-off must be assessed for each dye.
- After the dye is added to the medium, stir on a hot-plate until the dye is evenly distributed (Figure 1B).
- Pipet 5 ml of the dyed medium into 35-mm Petri dishes. You will be able to make 5 plates in total (Figure 1C).

Figure 1. Production of NG agar plates for staining of live C. elegans. A. NG medium after autoclaving but before addition of LysoTracker Red DND-99. B. NG medium after addition of LysoTracker Red DND-99. Note the pink color. C. 35-mm Petri dishes each with 5 ml LysoTracker-containing NG agar. - Put plates in a foiled container and dry at room temperature for at least 2 days. If plates are needed sooner, they can be dried in a hood (with the light off).
- Streak out E. coli OP50 on to an LB agar plate without antibiotics. It is important to streak to single colonies to ensure purity. Grow at 37 °C overnight.
- The next day, check the LB agar plate for OP50 colonies and possible contaminants. If OP50 colonies, but no contaminants, have grown, pick a single OP50 colony and inoculate a 5 ml LB liquid culture. Incubate with shaking at 37 °C overnight.
- Overnight growth will produce a saturated OP50 culture (~108-109 CPU/ml). Transfer 500 μl of the saturated OP50 culture to a microcentrifuge tube and add the same dye that was previously added to the NG agar plates (Figure 2A). Use the same final concentration of the dye as before.
- Aliquot 100 μl of the dyed OP50 culture on to the center of the dyed NG agar plates (Figure 2B). Keep plates in a foiled container at room temperature for at least 2 days. Bacteria will form a lawn. Plates can be maintained in a covered container for up to 1 week at room temperature, and are suitable for use as long as they do not dry out and crack.
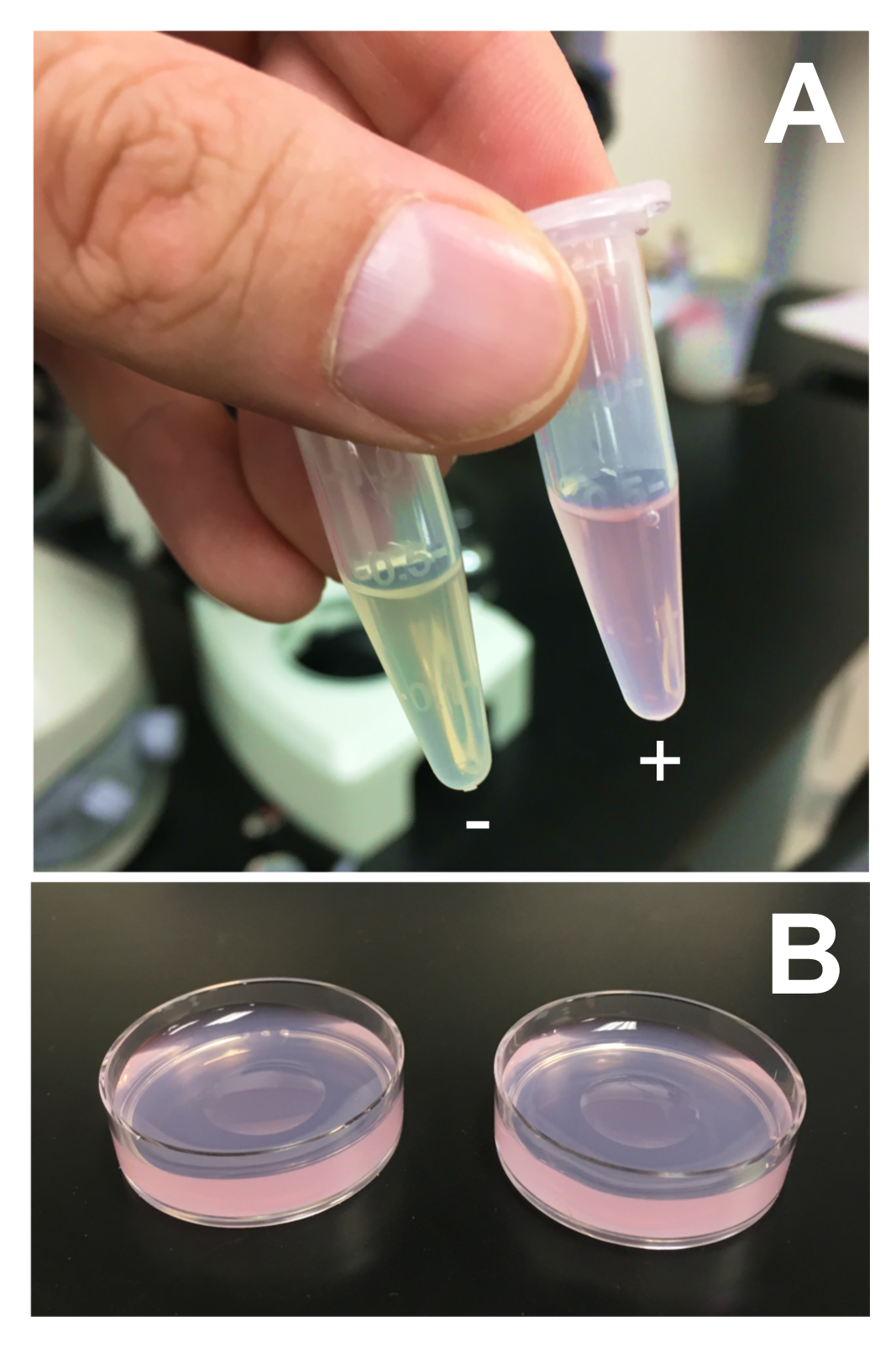
Figure 2. Preparation of dyed bacteria. A. OP50 aliquots with (+) or without (-) LysoTracker Red DND-99. B. Two LysoTracker plates with 100 μl OP50 spotted in the center. - For most robust staining, transfer worms to the bacterial lawns as eggs or L1 larvae. C. elegans eggs can be isolated by bleaching. For bleaching, gravid adult worms should be collected. Rinse plates with M9 buffer to collect the worms, and transfer the worms into a 15 ml Falcon tube. Centrifuge at 200 x g for 30 sec to pellet the worms. Remove the M9 buffer, and add 5 ml of fresh worm bleaching solution. Vortex for 5 min to dissolve the animals and release the eggs. Add 5 ml M9 buffer and invert the tube to mix. Pellet the released eggs at 200 x g for 30 sec, and wash the eggs three times with 5 ml M9 buffer. Plate the eggs directly, or nutate the eggs overnight in M9 buffer to allow the worms to develop into L1 larvae before plating. Either approach is suitable for this application. Introduce about 50 eggs/animals per plate; if more worms are included, the bacterial food source may be exhausted before worms reach adulthood.
- Put the foiled container with the worm plates into an appropriate incubator. For standard staining, incubation at 20 °C is suitable. When expression of germline transgenes is also desired, incubate at 25 °C to prevent germline silencing (Merritt et al., 2010). Allow the worms to develop into young adults (from the embryo stage, this will take around 3 days, depending on the incubation temperature).
- Make 1 mm-thick 4% agarose pads that can be used to mount the worms for microscopy. First, separate the two halves of an empty 1 mm gel cassette. Then, melt 1 g of agarose in 25 ml H2O in a glass beaker or flask. Before the agarose solidifies, pour the agarose on to the flat inside surface of one of the cassette halves, and compress the agarose to 1 mm thickness using the other cassette half. Once the agarose solidifies, pull the two halves of the cassette apart; a 1 mm-thick sheet of agarose should stick to one half of the cassette. Use a razor to cut the agarose into small square pads.
- Place an agarose pad onto a microscope slide and add a drop of 2 mM levamisole (a paralyzing agent used to immobilize worms). Transfer young-adult worms to the levamisole drop and place a coverslip on top.
- Secure the slide to the stage of a confocal microscope. Use appropriate filter sets to image the fluorophore of interest (Figure 3).
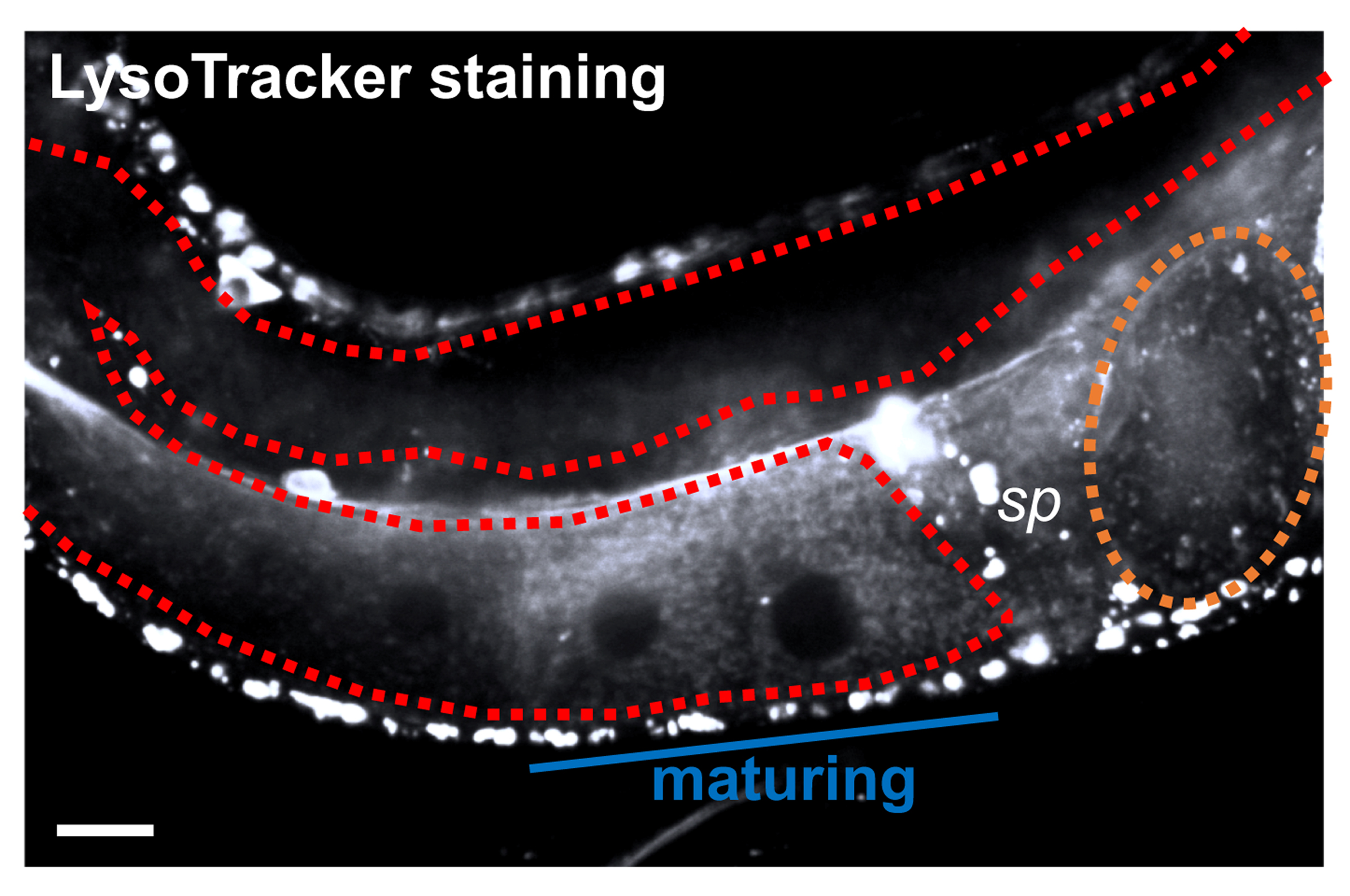
Figure 3. Example staining of lysosomes in the C. elegans germline. A gonad arm is outlined in red, and an embryo is outlined in orange. LysoTracker-positive puncta develop in maturing oocytes as they approach sperm (sp). Note that other tissues are also stained. Scale bar = 10 μm.
Data analysis
Staining patterns in the germline can be observed upon image acquisition (Figure 3). Fluorescence intensities can be quantified using ImageJ. In general, three independent replicates should be performed for quantification. Statistical analysis of staining patterns/intensities among experimental groups can be performed using unpaired t-tests, with significance defined as P < 0.05. Detailed information on data analysis and raw data for different types of experiments can be found in the original research article (Bohnert and Kenyon, 2017), which is freely available on PubMed Central (PMCID: PMC5936623).
Notes
It is important to note that tissues other than the germline will also be stained (Figure 3). Because worms are being fed the dye, the intestine will tend to fluoresce brightly when in focus. It is easiest to image full gonad arms in live animals when proximal and distal regions of the germline are on the same z-plane and are situated on top of the intestine.
This staining approach can be applied to label the germlines of animals treated with RNAi. In this case, the dye would need to be added to RNAi plates (NG agar supplemented with 100 μg/ml carbenicillin and 1 mM IPTG) and to the relevant bacterial strains (i.e., those expressing RNAi clones). When gene knockdown is desired in late development or adulthood, worms should first be grown on normal NG plates containing the dye, and then moved to dyed RNAi plates at the appropriate stage.
Recipes
- 1 M CaCl2
Dissolve 147.0 g CaCl2•2H2O in 1 L H2O, and autoclave using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min - 5 mg/ml cholesterol
Dissolve 0.25 g cholesterol in 50 ml 200-proof ethanol, and filter-sterilize - 1 M MgSO4
Dissolve 120.4 g MgSO4 in 1 L H2O, and autoclave using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min - 1 M KPO4 pH 6.0
Dissolve 108.3 g KH2PO4 and 35.6 g K2HPO4 in 1 L H2O, and autoclave using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min - 100 mg/ml carbenicillin
Dissolve 1 g carbenicillin in 10 ml H2O, and filter-sterilize - 1 M IPTG
Dissolve 2.38 g IPTG in 10 ml H2O, and filter-sterilize - NG agar
- Mix 75 mg NaCl, 62.5 mg peptone, and 425 mg agar in 25 ml H2O
- Autoclave using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min
- Cool at room temperature until the flask can be comfortably touched
- Add 25 μl 1 M CaCl2, 25 μl 5 mg/ml cholesterol, 25 μl 1 M MgSO4, and 625 μl 1 M KPO4 pH 6.0
- Then, proceed to adding the desired dye
- Dye stock solutions
Make a 1 mM stock solution of each dye in DMSO (or other recommended solvent).
Notes:- These stock solutions can be used at 1:500 to give a 2 μM final concentration. As noted in the Procedure, different concentrations may be suitable for other dyes that have yet to be verified.
- Stocks should be protected from light as much as possible (for example, tubes can be wrapped in foil and maintained within boxes). Recommended storage of liquid stocks is -20 °C.
- LB medium
- Dissolve 25 g LB Broth (Miller) in 1 L H2O, and autoclave using a liquid cycle set at 121 °C and 15 psi pressure for at least 30 min
- To make LB agar, add 8 g agar per 400 ml LB before autoclaving
- LB agar can be poured into 100-mm Petri dishes
- Worm bleaching solution
Mix 3.5 ml H2O, 1 ml household bleach, and 0.5 ml 5 N NaOH (Stiernagle, 2006)
To make 5 N NaOH: dissolve 10 g NaOH in 50 ml H2O - Agarose pads
- Add 1 g agarose to 25 ml H2O in a beaker
- Microwave to dissolve the agarose
- Pour agarose on to a clean, inside surface of an old protein-gel cassette and compress to ~1 mm in thickness using the cassette’s other half
- Cut agarose squares roughly 1.25 cm x 1.25 cm using a razor (the exact size and geometry of the pads is not a strict requirement)
- Levamisole solution
- Make a 1 M stock solution by dissolving 0.24 g levamisole hydrochloride in 1 ml H2O
- Dilute 1:500 in H2O to make a 2 mM working solution
Acknowledgments
The study (Bohnert and Kenyon, 2017) on which this full protocol is based was originally performed in Cynthia Kenyon’s lab at UCSF and Calico Life Sciences. I thank Cynthia for her mentorship, generosity, and creative insight, and members of the Kenyon laboratory for helpful comments in designing these experiments. I also thank Maria Ingaramo and Andy York for sharing microscopy advice and expertise. While in the Kenyon lab, I was an Honorary Fellow of the Jane Coffin Childs Memorial Fund. I am now an Assistant Professor at Louisiana State University. I thank my current lab members and Alyssa Johnson for thought-provoking discussions and input on this protocol.
Competing interests
I declare no conflicts of interest or competing interests.
References
- Bohnert, K. A. and Kenyon, C. (2017). A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature 551(7682): 629-633.
- Hermann, G. J., Schroeder, L. K., Hieb, C. A., Kershner, A. M., Rabbitts, B. M., Fonarev, P., Grant, B. D. and Priess, J. R. (2005). Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell 16(7): 3273-3288.
- Klass, M. R. (1977). Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev 6(6): 413-429.
- Merritt, C., Gallo, C. M., Rasoloson, D., and Seydoux, G. (2010). Transgenic solutions for the germline. WormBook
- Page, A. and Johnstone, I. L. (2007). The cuticle. WormBook
- Schaheen, L., Dang, H. and Fares, H. (2006). Basis of lethality in C. elegans lacking CUP-5, the mucolipidosis type IV orthologue. Dev Biol 293(2): 382-391.
- Stiernagle, T. (2006). Maintenance of C. elegans. WormBook
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bohnert, K. A. (2018). Staining the Germline in Live Caenorhabditis elegans: Overcoming Challenges by Applying a Fluorescent-dye Feeding Strategy. Bio-protocol 8(21): e3077. DOI: 10.21769/BioProtoc.3077.
Category
Cell Biology > Cell imaging > Confocal microscopy
Cell Biology > Cell staining > Germline
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










