- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
mRNA Stability Assay Using Transcription Inhibition by Actinomycin D in Mouse Pluripotent Stem Cells
Published: Vol 8, Iss 21, Nov 5, 2018 DOI: 10.21769/BioProtoc.3072 Views: 37567
Reviewed by: Xingrong DuHui Chen WuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Automated 384-well SYBR Green Expression Array for Optimization of Human Induced Pluripotent Stem Cell Differentiation
Max Y. Chen [...] Birgitt Schuele
Jun 5, 2023 2239 Views

Absolute Quantification of mRNA Isoforms in Adult Stem Cells Using Microfluidic Digital PCR
Shubhangi Das Barman [...] Antoine de Morree
Sep 5, 2023 1853 Views

Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Ruben Warkentin and Anna Marie Pyle
Oct 20, 2025 1393 Views
Abstract
Gene expression is regulated through multiple steps at both transcriptional and post-transcriptional levels. The net abundance of mature mRNA species in cells is determined by the balance between transcription and degradation. Thus, the regulation of mRNA stability is a key post-transcriptional event that can greatly affect the net level of mRNAs in cells. The mRNA stability within cells can be measured indirectly by analyzing the mRNA half-life following transcription inhibition, where changes in mRNA levels are assumed to reflect mRNA degradation. Determination of mRNA half-life as a measure of mRNA stability is useful in understanding gene expression changes and underlying mechanisms regulating the level of transcripts at different physiological conditions or developmental stages. The protocol described here presents the analysis of mRNA decay as a measure for determining mRNA stability after transcriptional inhibition with Actinomycin D treatment in control and SRSF3 depleted mouse induced pluripotent stem cells (iPSC).
Keywords: mRNA stabilityBackground
Determining the stability of mRNA within cells provides an important measure for understanding post-transcriptional gene regulation and the potential role of RNA-protein interactions in the process. Under any specific condition such as following extracellular stimuli or gene knockdown the stability of mRNAs may change due to enhanced degradation or extended half-lives (Shyu et al., 1989). Therefore, to assess mRNA stability, direct measurements of decay rates of endogenous mRNAs have been performed in a number of ways, including kinetic labeling techniques and the use of transcriptional inhibitors (Chen et al., 2008). One of the simplest techniques of measuring mRNA stability is by inhibiting transcription in vivo with transcription inhibitors and measuring the mRNA kinetics.
Actinomycin D is a transcription inhibitor which intercalates into DNA. Actinomycin D forms a very stable complex with DNA, preventing the unwinding of the DNA double-helix, thus inhibiting the DNA-dependent RNA polymerase activity. Actinomycin D is widely used in mRNA stability assays to inhibit the synthesis of new mRNA, allowing the assessment of mRNA decay by measuring mRNA abundance following transcription inhibition (Avendano and Menéndez, 2008). At low concentrations, Actinomycin D inhibits transcription without significantly affecting DNA replication or protein synthesis (Berg et al., 2002). Other transcription inhibitors such as 5,6-dichloro-1β-1-ribofuranosylbenzimidazole (DRB) which interacts directly with the RNA polymerase II have also been successfully used in similar assays (Harrold et al., 1991). Please refer to Bensaude (2011) for more detailed information on different transcription inhibitors that can be used and their specific properties. More advanced techniques such as the use of inducible promoters to control transient transcription have presented advantages over the potential cytotoxic effects of Actinomycin D or other transcription inhibitors in the analysis of mRNA decay (Chen et al., 2008). However, the advantage of Actinomycin D assay is that it does not require the construction and introduction of exogenous genes into cells, and provides a way of measuring stability changes of endogenous mRNAs (Chen et al., 2008).
We have established serine-arginine-rich splicing factor 3 (SRSF3)-RNA interactions as a critical means to co-ordinate gene expression in pluripotent cells (Ratnadiwakara et al., 2018). Further, SRSF3 has been reported to regulate mRNA levels including its own mRNA abundance via alternative splicing coupled to nonsense mediated decay (NMD) (Anko et al., 2012). To determine if SRSF3 affects the production or stability of NMD-sensitive transcript variants in pluripotent cells, we determined mRNA half-lives of SRSF3 target mRNAs in Actinomycin D treated control and SRSF3-depleted pluripotent stem cells (Ratnadiwakara et al., 2018). Several techniques such as Northern blot analysis, in situ hybridization and quantitative PCR can be used to determine the mRNA half-life after transcription inhibition. We used quantitative PCR which allows rapid and sensitive measurement of half-lives of mRNAs across a broad range of expression levels, including low abundant mRNAs. The protocol described here can be used to successfully measure mRNA decay in pluripotent stem cells.
Materials and Reagents
- 6-well cell culture plates (Sigma-Aldrich, catalog number: CLS3516)
- Serological pipettes 10 ml (Sigma-Aldrich, catalog number: CLS4488)
- 15 ml Falcon tubes (Sigma-Aldrich, catalog number: CLS430791)
- Sterile filter pipette tips 10 µl, 20 µl, 200 µl, 1,000 µl (Axygen, catalog numbers: TF10LRS, TF20LRS, TF200LRS and TF1000LRS)
- Microcentrifuge tubes (Axygen, catalog number: MCT-175-C)
- Neptune semi-skirted 96-well plates (VWR, catalog number: 89126-694)
- Optically clear adhesive seal sheets (Thermo Fisher Scientific, catalog number: AB-1170)
- Knock-Out DMEM (Thermo Fisher Scientific, catalog number: 10829018)
- ES cell grade fetal bovine serum (Sigma-Aldrich, catalog number: F9423)
- GlutaMAX (Life Technologies, catalog number: 35050-061)
- Trypsin 0.25% (Life Technologies, catalog number: 25200-056)
- Penicillin-Streptomycin (Life Technologies, catalog number: 15070-063)
- Non-Essential Amino Acids (Life Technologies, catalog number: 11140-050)
- Beta-Mercapto Ethanol (Life-technologies, catalog number: 21985-023)
- Leukaemia inhibitory factor (LIF) (here LIF was produced by the Monash University Protein Production Unit, Australia)
- Phosphate-buffered saline (PBS) (Life Technologies, catalog number: 14190-250)
- Cells in culture (here mouse induced pluripotent stem cells)
- (z)-4-Hydroxytamoxifen (Sigma-Aldrich, catalog number: H7904-5MG)
- Cell culture grade Actinomycin D (Sigma-Aldrich, catalog number: A9415)
- Cell culture grade Dimethyl Sulfoxide (DMSO) (AppliChem, catalog number: A3672,0100)
- TRI Reagent (Sigma-Aldrich, catalog number: T9424)
- Chloroform (Sigma-Aldrich, catalog number: 288306)
- Isopropanol (Sigma-Aldrich, catalog number: 278475)
- RNA grade Glycogen (Thermo Fisher Scientific, catalog number: R0551)
- Ethanol (any molecular grade)
- RNase-free water (Invitrogen, catalog number: 10977-015)
- RQ1 DNase kit (Promega, catalog number: M6101)
- SuperScript III Reverse transcription kit (Thermo Fisher Scientific, catalog number: 18080044)
- RNaseOUT (Thermo Fisher Scientific, catalog number: 10777019)
- Random hexamer primer mix (Bioline, catalog number: BIIO38028)
- OligodT18 (IDT)
- SYBR green master mix, here Luminaries HiGreen qPCR Master Mix, Low ROX (Thermo Fisher Scientific, catalog number: K0974)
- qPCR primers for the genes of interest
- ES culture media (see Recipes)
Equipment
- Sterile cell culture hood (Safemate Vision 1.2 cabinet, catalog number: LDE0820)
- 37 °C cell culture incubator with 10% CO2 and 5% O2 (hypoxia) (Thermo Fisher Scientific, model: HeracellTM 150)
- Automated cell counter (NanoEnTek, catalog number: E1000)
- Refrigerated microcentrifuge (Bio-strategy, catalog number: 75002421)
- qPCR machine (7500 Real-Time PCR System) (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4351105)
- Vortexer
- Freezer
Software
- SDSv2.4 (Thermo Fisher Scientific, www.thermofisher.com/au/en/home/technical-resources/software-downloads/applied-biosystems-7900ht-fast-real-timespcr-system.html)
- GraphPad Prism 7 (GraphPad Sowtware, Inc, www.graphpad.com)
- Microsoft Excel Version 15.41 (Microsoft)
Procedure
- Cell culture and sample generation
- Seed 3 x 105 cells per well in 3 ml of media in each well of a 6-well plate (Figure 1–step 1, in total 6 wells per replicate).
Note: We used mouse induced pluripotent stem cells (iPS cells) generated from reprogrammable tamoxifen inducible SRSF3-knockout and control mouse embryonic fibroblasts (MEFs) (Ratnadiwakara et al., 2018). The culture conditions described here reflect the requirements for these cells. A detailed description of the generation and culture properties of the iPS cell lines used here can be found in (Ratnadiwakara et al., 2018) and the media composition can be found under Recipes. This protocol is applicable to a wide range of tissue culture cell lines and the culture properties should be adjusted depending on the cell line used. - Let the cells adhere to the culture dish for 4 h, after which treat the cells with 5 µM tamoxifen (4OHT) to induce Cre-activity and SRSF3 depletion (Figure 1–step 2).
Note: We have tested a range of tamoxifen (4OHT) concentrations and 5 µM 4OHT results in high recombination efficiency with minimal cytotoxicity. - After 24 h, collect the cells from the first well as the first-time point by brief trypsination (0.1% trypsin for 3 min at 37 °C) or by using a cell scraper; t = 0 (Figure 1–step 3). A rapid processing of samples is required for the accuracy of the time points.
- Spin down the collected cells at 470 x g for 3 min at 20 °C.
- Re-suspend the cell pellet in 1 ml of TRI Reagent and freeze at -80 °C.
- To the remaining 5 wells, add 30 µl of 1 mg/ml Actinomycin D stock to obtain a final concentration of 10 µg/ml in 3 ml of culture media (Figure 1–step 3).
Note: Make 1 mg/ml Actinomycin D stock in DMSO and freeze in aliquots at -20 °C. Dilute 30 µl of Actinomycin D stock in 100 µl of media and add dropwise to each well for uniform distribution. - Collect samples at 1, 2, 4, 6 and 8 h time points following Actinomycin D addition and freeze the cell pellet in 1 ml of TRI Reagent as described above (Figure 1–steps 4 and 5).
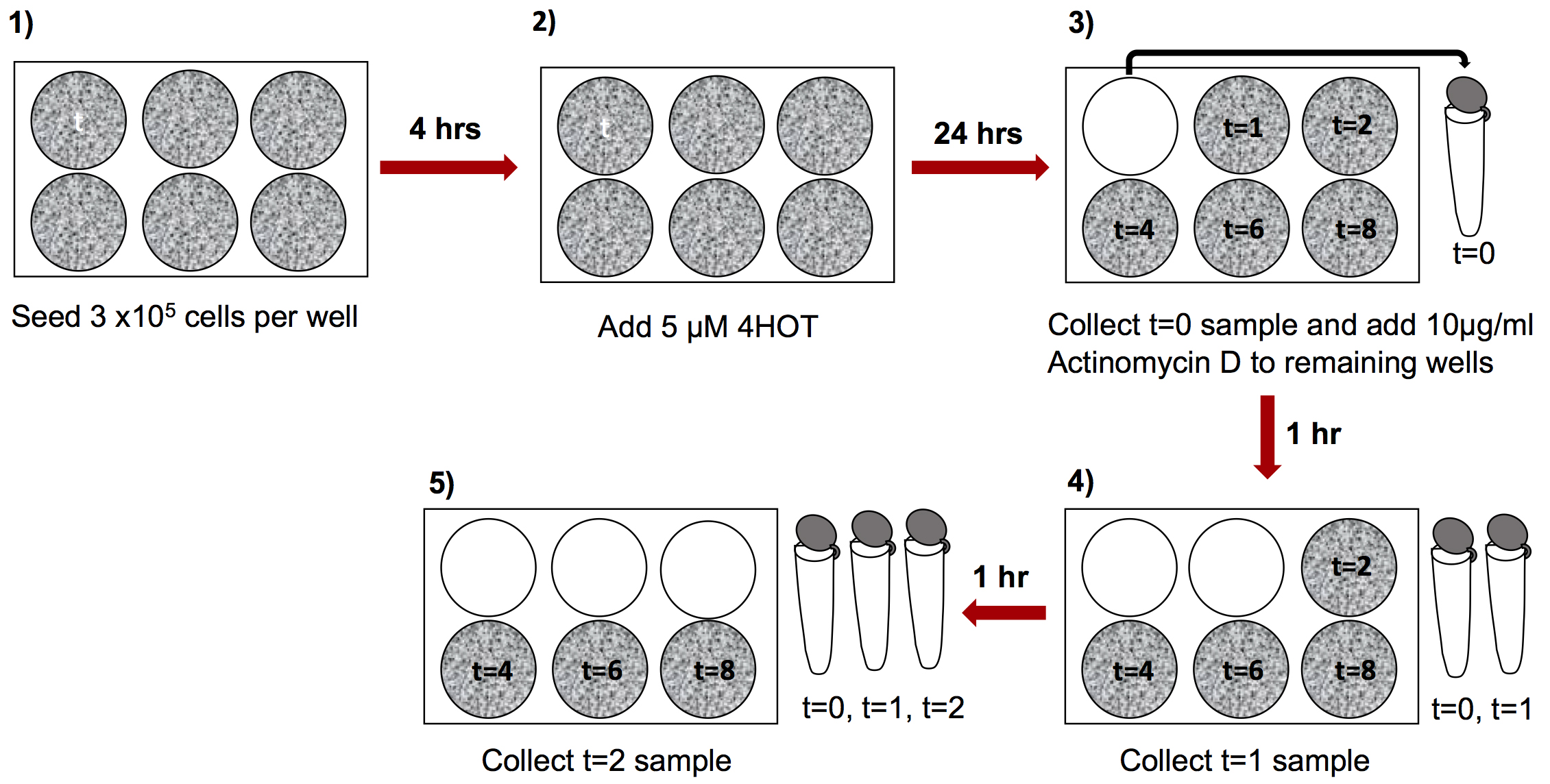
Figure 1. Experimental setup and sample collection. Collect the samples at relevant time points and proceed to RNA extraction.
- Seed 3 x 105 cells per well in 3 ml of media in each well of a 6-well plate (Figure 1–step 1, in total 6 wells per replicate).
- RNA extraction
- Thaw the cells frozen in TRI Reagent at room temperature for 10 min.
- Add 200 µl of chloroform and shake vigorously for 15 s and allow to stand for 10 min at room temperature.
- Centrifuge at 12,000 x g for 15 min at 4 °C.
- Transfer the aqueous layer into a fresh tube and add 500 µl of 2-propanol and 1 µl of Glycogen.
Note: Addition of glycogen helps the maximum recovery of the RNA and visualization of the RNA pellet. - Mix carefully and allow to stand at room temperature for 10 min to precipitate the RNA.
- Centrifuge the mixture at 12,000 x g for 10 min at 4 °C.
- Remove the supernatant and wash the RNA pellet by adding 1 ml of 75% ethanol, vortex and centrifuge at 7,500 x g for 5 min at 4 °C.
- Briefly air-dry the RNA pellet for 5-10 min.
Note: Avoid over-drying the pellet as this will greatly decrease its solubility. - Re-suspend RNA in 30 µl of RNase-free water and proceed to cDNA Synthesis.
- Reverse transcription and quantitative real-time PCR
- Perform DNaseI treatment on all samples to remove potential genomic DNA contamination that can affect the downstream analysis. We used Promega RQ1 DNase kit according to the manufacturer’s protocol and use 1 µg of RNA per sample.
- Carry out the reverse transcription of the RNA samples using a reverse transcription kit. We used SuperScript III Reverse transcriptase according to the manufacture’s protocol with equal amounts of random hexamers and oligodT18 for reverse priming. In short, 1 µg of DNaseI-treated RNA is incubated with dNTPs and primers at 65 °C for 5 min, followed by the addition of SuperScript III reagents. The cDNA synthesis is performed at 50 °C for 1 h and the reaction terminated by incubation at 70 °C for 15 min.
Note: A mixture of random hexamers and oligodT18 can improve the sensitivity of the cDNA synthesis. - Dilute the cDNA 1:10 with nuclease-free water to be used as a template for quantitative PCR.
- Perform quantitative PCR with the primers specific for the gene of interest. Use the optimized manufacturer’s protocol for specific SYBR Green master mixes. We used 5 µl of Luminaries 2x HiGreen qPCR Master Mix per reaction, optimized concentration of each forward and reverse primers (typically 0.15-0.30 µM end concentration) and 2 µl of diluted cDNA per reaction. Perform two or more technical replicates for each sample.
Data analysis
- Upon the completion of the PCR, run the machine specific software to extract the data. We used SDS software.
Note: The dissociation curve should produce a single peak for each reaction indicating the amplification of a single specific product (Figure 2C). - Export Ct values to Excel spread sheet and calculate the average of replicates for each reaction.
- Normalize the Ct average of each time point to the Ct average value of t = 0 to obtain ∆Ct value.
∆Ct = (Average Ct of each time point - Average Ct of t=0). - Calculate the relative abundance for each time point.
mRNA abundance = 2(-∆CT) - Plot the relative abundance of mRNA at each time point relative to t = 0 using GraphPad Prism or similar software (Figures 2A and 2D).
- Determine the mRNA decay rate by non-linear regression curve fitting (one phase decay) using GraphPad Prism (Figure 2B). We used following parameters:
- Least squares (ordinary fit)
- Confidence level–95%
- Asymmetrical (likelihood) CI
- Goodness of fit was quantified with R square
- Convergence criteria–medium
- This protocol describes one biological replicate but at least three independent experiments should be performed for statistical assessment.
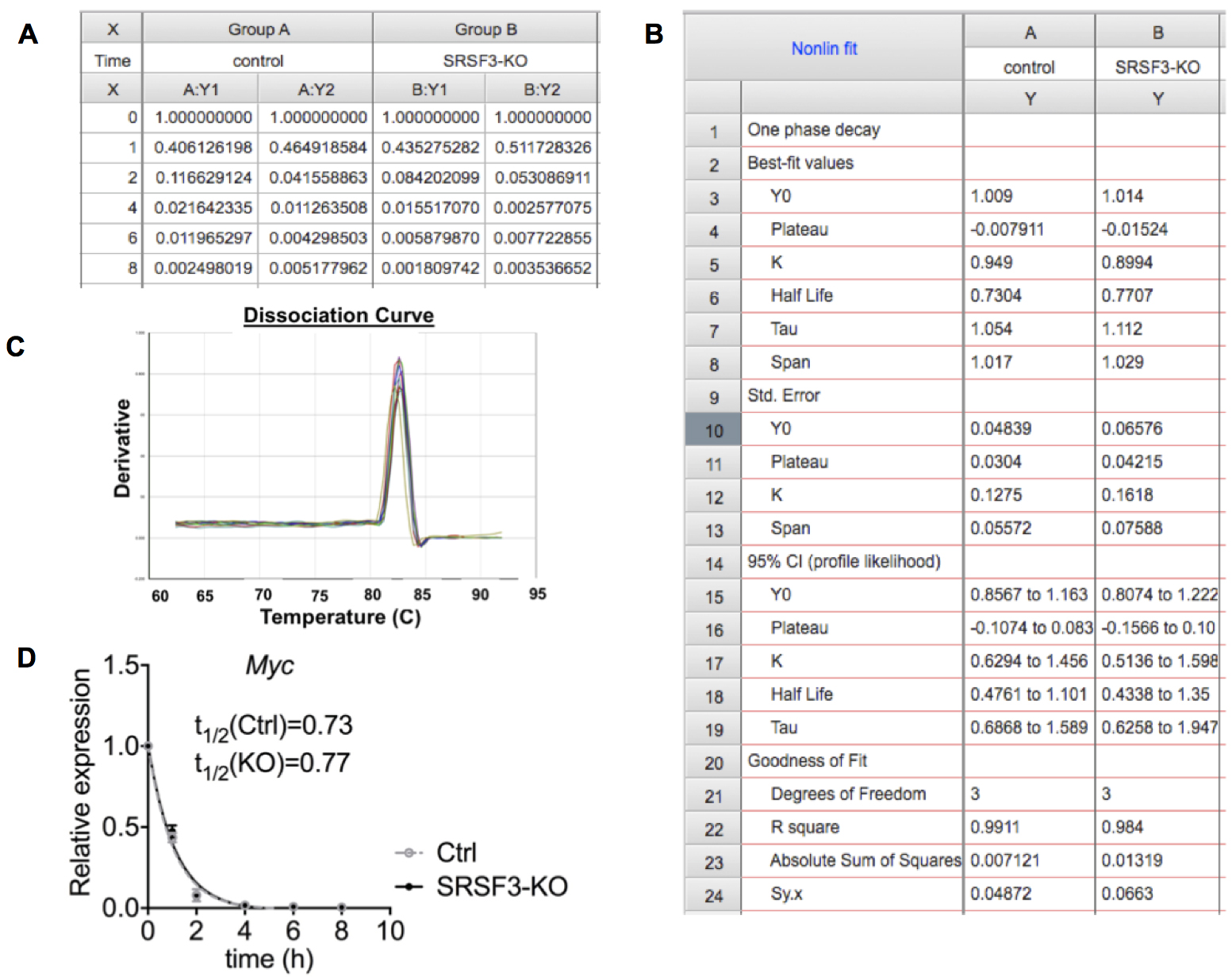
Figure 2. Data analysis. A. Relative Myc mRNA abundance (2(-∆CT)) for each time point for 2 replicates in control and SRSF3 depleted iPS cell samples. B. One phase decay analysis in GraphPad Prism. C. The dissociation curve with a single peak indicating the amplification of a single PCR product. D. Graph representing Myc mRNA decay in control and SRSF3-knockout (KO) cells after Actinomycin D treatment demonstrating similar half-lives for Myc mRNA in control and SRSF3-KO cells.
Recipes
- ES culture media
1x Knock-Out DMEM
15% ESC grade fetal bovine serum
2 mM GlutaMAX
1 mM Non-essential amino acids
1x Penicillin-streptomycin
Add 1,000 U/ml Leukemia Inhibitory Factor (LIF) and 1 µM beta-mercaptoethanol just before use
Acknowledgments
MLA was supported by National Health and Medical Research Council (NHMRC) GNT1043092 and GNT1138870, Aatos and Jane Erkko Foundation and Monash Biomedicine Discovery Fellowship.
Competing interests
The authors have no conflicts of interest or competing interests.
References
- Anko, M. L., Muller-McNicoll, M., Brandl, H., Curk, T., Gorup, C., Henry, I., Ule, J. and Neugebauer, K. M. (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13(3): R17.
- Avendano, C. and J. Menéndez (2008). DNA Intercalators and topoisomerase inhibitors. Medicinal Chemistry of Anticancer Drugs. Elsevier Inc.: Madrid, Spain: 199-228.
- Bensaude, O. (2011). Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2(3): 103-108.
- Berg J. M., Tymoczko J. L. and Stryer L. (2002). Transcription Is Catalyzed by RNA Polymerase. In: Biochemistry. 5th edition. New York: W H Freeman and Company.
- Chen, C. Y., Ezzeddine, N. and Shyu, A. B. (2008). Messenger RNA half-life measurements in mammalian cells. Methods Enzymol 448: 335-357.
- Harrold, S., Genovese, C., Kobrin, B., Morrison, S. L. and Milcarek, C. (1991). A comparison of apparent mRNA half-life using kinetic labeling techniques vs decay following administration of transcriptional inhibitors. Anal Biochem 198(1): 19-29.
- Ratnadiwakara, M., Archer, S. K., Dent, C. I., Ruiz De Los Mozos, I., Beilharz, T. H., Knaupp, A. S., Nefzger, C. M., Polo, J. M. and Anko, M. L. (2018). SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife 7: e37419.
- Shyu, A. B., Greenberg, M. E. and Belasco, J. G. (1989). The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev 3(1): 60-72.
Article Information
Copyright
Ratnadiwakara and Änkö. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ratnadiwakara, M. and Änkö, M. (2018). mRNA Stability Assay Using Transcription Inhibition by Actinomycin D in Mouse Pluripotent Stem Cells. Bio-protocol 8(21): e3072. DOI: 10.21769/BioProtoc.3072.
- Ratnadiwakara, M., Archer, S. K., Dent, C. I., Ruiz De Los Mozos, I., Beilharz, T. H., Knaupp, A. S., Nefzger, C. M., Polo, J. M. and Anko, M. L. (2018). SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. Elife 7: e37419.
Category
Molecular Biology > RNA > RNA detection
Stem Cell > Pluripotent stem cell > Cell pluripotency
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








