- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microtitre Plate Based Cell-SELEX Method
(*contributed equally to this work) Published: Vol 8, Iss 20, Oct 20, 2018 DOI: 10.21769/BioProtoc.3051 Views: 7961
Reviewed by: Longping Victor TseVaibhav B. ShahAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fluorescence Dequenching-based Liposome Leakage Assay to Measure Membrane Permeabilization by Pore-forming Proteins
Javier Aguilera [...] Jianjun Sun
May 20, 2021 7113 Views

A Gel-Based Assay for Probing Protein Translocation on dsDNA
Christiane Brugger and Alexandra M. Deaconescu
Jul 20, 2021 4337 Views

Extraction and Electrophoretic Analysis of Bacterial Lipopolysaccharides and Outer Membrane Proteins
Yue-Jia Lee and Thomas J. Inzana
Dec 20, 2021 5750 Views
Abstract
Aptamers have emerged as a novel category in the field of bioreceptors due to their wide applications ranging from biosensing to therapeutics. Several variations of their screening process, called SELEX have been reported which can yield sequences with desired properties needed for their final use. We report a facile microtiter plate-based Cell-SELEX method for a gram-negative bacteria E. coli. The optimized protocol allows the reduction of number of rounds for SELEX by offering higher surface area and longer retention times. In addition, this protocol can be modified for other prokaryotic and eukaryotic cells, and glycan moieties as target for generation of high affinity bio-receptors in a short course of time in-vitro.
Keywords: AptamersBackground
Aptamers are single strands of synthetic DNA or RNA described in 1990 (Ellington and Szostak, 1990; Tuerk and Gold, 1990) with a distinct 3D geometry, which is a manifestation of their sequences. Different sequences allow the synthesis of aptamers, which can bind to an array of molecules ranging from small molecules to large proteins. This allowed aptamers to emerge as the rivals to conventional antibodies, which are limited to proteins as their targets due to structural limitations and can not be raised against high-risk pathogens as these usually kill the host much earlier than the time required to generate high affinity antibodies. Aptamers are generated in in-vitro setups and thus can effectively be used for high-risk pathogens. SELEX (Systematic Evolution of Ligands by EXponential enrichment) is an iterative process involving repeated exposures and separation leading to screening of a pool with dissociation constants in low nM ranges. Several variations of SELEX have been proposed which use a variety of stationary matrices including capillary electrophoresis SELEX, affinity chromatography based SELEX, magnetic bead SELEX, in-vivo SELEX, FACS SELEX and Microfluidics based SELEX, etc. These methods present various advantages over others but the number of SELEX rounds are consistently high.
A standard SELEX method has four major steps; exposure of random sequence single stranded nucleotide oligo library to target, binding of oligos to target molecule, selection of binders and removal of non-binding oligos, amplification of the binder fraction and portioning of the amplicon to single strand. These steps are iteratively performed till pool of high binding aptamer are screened out from the library. The oligonucleotide library consists of a random base-sequence flanked on both ends by primer binding sites, which aids in amplification and enrichment (Safeh et al., 2010; Kim et al., 2013).
We report a microtiter plate-based Cell-SELEX method, which can be employed over a wide number of molecules and cell types (Figure 1). The advantage proposed by the method is uniquely low number of rounds, which are the outcome of large surface area, higher retention times and a novel superior bio-probe based partitioning process (Priyanka et al., 2014).
Figure 1. Concise Protocol Schematic. A step-by-step flowchart showing the various steps of Cell-SELEX. (Modified from author’s work Kaur et al., 2017)
Materials and Reagents
- Pipette tips, 10 μl (Tarsons, catalog number: 523100 )
- Pipette tips, 200 μl (Tarsons, catalog number: 523101 )
- Pipette tips, 1 ml (Tarsons, catalog number: 523104 )
- Nunc Maxisorp F96 microtitre plates (Thermo Fisher Scientific, catalog number: 437111 )
- Centrifuge tubes 50 ml (Tarsons, catalog number: 546041 )
- Microcentrifuge tubes (MCT) 1.5 ml (Tarsons, catalog number: 500010 )
- PCR Tubes 0.2 ml (Tarsons, catalog number: 510051 )
- Bacterial strains (E. coli)
- ssDNA library and primers (IDT custom synthesis):
Forward Primer: 5'-ATCCAGAGTGACGCAGCA-3'
Reverse Primer: 5'-biotin-ACTAAGCCACCGTGTCCA-3'
Library: 5'-ATCCAGAGTGACGCAGCA-45N-TGGACACGGTGGCTTAGT-3' - LB broth (HiMedia Laboratories, catalog number: M1245 )
- Phenylboronic acid (HiMedia Laboratories, catalog number: RM1599 )
- Streptavidin-Gold from Streptomyces avidinii (Sigma-Aldrich, catalog number: 53134 )
- Tris-HCl (HiMedia Laboratories, catalog number: MB030 )
- Glycine (HiMedia Laboratories, catalog number: MB013 )
- Magnesium chloride (HiMedia Laboratories, catalog number: MB040 )
- Sodium chloride (HiMedia Laboratories, catalog number: MB023 )
- Sodium phosphate monobasic (HiMedia Laboratories, catalog number: GRM3963 )
- Sodium phosphate dibasic (HiMedia Laboratories, catalog number: MB024 )
- Tris base (HiMedia Laboratories, catalog number: TC072 )
- Sodium carbonate (HiMedia Laboratories, catalog number: GRM851 )
- Sodium bicarbonate (HiMedia Laboratories, catalog number: TC230 )
- Hydrochloric acid 37% (Merck, catalog number: 100317 )
- PCR mastermix (2x) (Thermo Fisher Scientific, catalog number: K0171 )
- PCR grade DMSO (Sigma-Aldrich, catalog number: D9170 )
- Tryptone (HiMedia Laboratories, catalog number: RM014 )
- Yeast extract powder (HiMedia Laboratories, catalog number: RM027 )
- LB growth media (see Recipes)
- Carbonate buffer (see Recipes)
- Tris-HCl binding buffer (see Recipes)
- Glycine-HCl elution buffer (see Recipes)
- Tris neutralization solution (see Recipes)
- Phenylboronic acid (PBA) coating solution (see Recipes)
- Phosphate buffered saline (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, model: FinnpipetteTM F1 variable volume single channel pipettes)
- Incubator-Shaker (Eppendorf, model: Innova® 44 )
- 4 °C refrigerator (Vestfrost Solutions, model: AKG 377 )
- -20 °C freezer (Vestfrost Solutions, model: CFS 344 )
- pH meter (Hanna Instruments, model: pH 211 )
- Spectrophotometer (GE Healthcare, model: NanoVue Plus )
- Centrifuge (Eppendorf, model: 5430 R )
- Thermocycler (Bio-Rad Laboratories, model: C1000 TouchTM )
Procedure
Day 0
- Preparation of inoculum
- Take the lyophilized E. coli cells and resuspend them in 1 ml of sterile LB broth.
- Incubate it at 37 °C overnight with constant shaking at 180 rpm in a Incubator-shaker.
- Microtiter plate functionalization
- Take a sterile microtiter plate and coat it with 100 μl of 2.5 mM PBA prepared in filter-sterile carbonate buffer (pH 9.2).
- Incubate it overnight at 4 °C for the well surface to functionalize with PBA.
- Naïve aptamer library preparation
- Add 10 μl of 100 μM ssDNA library to 190 μl of binding buffer.
- Incubate it at 95 °C for 10 min and transfer to ice-bath immediately to prevent rehybridization of the single stranded DNA library.
Note: Step C2 can be repeated by reheating the DNA sample if user fails to transfer the materials to ice-bath. If heating on a heat-block with cooling assembly, use of cooling assembly to chill the samples should be avoided. - Transfer this to 4 °C refrigerator to incubate overnight for the single stranded DNA oligos to take their respective 3D conformations.
Day 1
- Growth and harvesting of cells
- Measure the growth of the inoculum using a spectrophotometer at λ = 600 nm. The optical density (O.D.) should be ≥ 1.
- Take 250 μl of inoculum and inoculate in 25 ml sterile LB broth.
Note: Use 1% of 1 O.D. cells as inoculum for subculturing. - Incubate it at 37 °C at 180 rpm for 1.5 h, till the E. coli cells reach log-phase (0.5-0.6 O.D.).
Note: For different cells, the growth curve can be measured prior to cell harvesting for SELEX rounds. - Centrifuge the culture at 3,000 x g for 10 min to form a soft pellet.
- Wash the cells thrice with phosphate buffered saline (PBS) (10 mM, pH 7.4) to remove residual media components.
Note: Washing involves resuspending pellet in PBS, mild vortexing and pelleting at 3,000 x g for 10 min. - Measure the O.D. using a spectrophotometer at λ = 600 nm. The optical density (O.D.) should be ≥ 1.
Note: 1 O.D. E. coli cells correspond to approximately 1 x 108 cells. Further dilution of cells can be made in PBS.
- SELEX INITIAL ROUND: Exposure and Screening
- Wash the PBA coated plate with carbonate buffer twice and resuspend 100 μl of the 1 x 103 cells in well 1.
- Incubate at 25 °C for 1 h, to allow the cells to coat the surface of the well.
- Decant the cell suspension by inverting the plate and wash the well with 200 µl of binding buffer thrice to remove any cells not bound to the well surface.
- Meanwhile, incubate 200 μl of prepared naïve library from Day 0 at 37 °C for 15 min.
- Gently add 200 μl of the prepared library to the well coated with cells.
- Incubate at 37 °C for 1 h.
- Decant the solution from the microtiter well by inverting the plate.
- Gently wash with 200 µl of binding buffer thrice to separate loosely bound sequences.
- Add 200 μl of elution buffer to the microtiter well.
- Incubate for 5 min at room temperature.
- Remove the solution from the well by using a micropipette carefully without touching the walls of the well and immediately neutralize by adding 10 μl of neutralization buffer to bring the pH to ~7.
- Quantify the obtained sequences using NanoVue spectrophotometer and label as Round 1.
Note: To avoid any bacterial contamination due to detachment, the DNA elutes can be centrifuged at 3,000 x g for 10 min and supernatant taken, prior to quantification. - The screened pool can be stored at -20 °C till further use.
Note: Discard the contents in Steps B3, B7 and B8. Contents in Step B11 is to be saved and proceeded with for the next steps.
Day 2
SELEX INITIAL ROUND: Amplification and Partitioning
- The Round 1 pool is further amplified to increase the concentration of the binding sequences. Carry the following steps on ice.
- To prepare the PCR reaction mixture (50 μl); mix the following components:
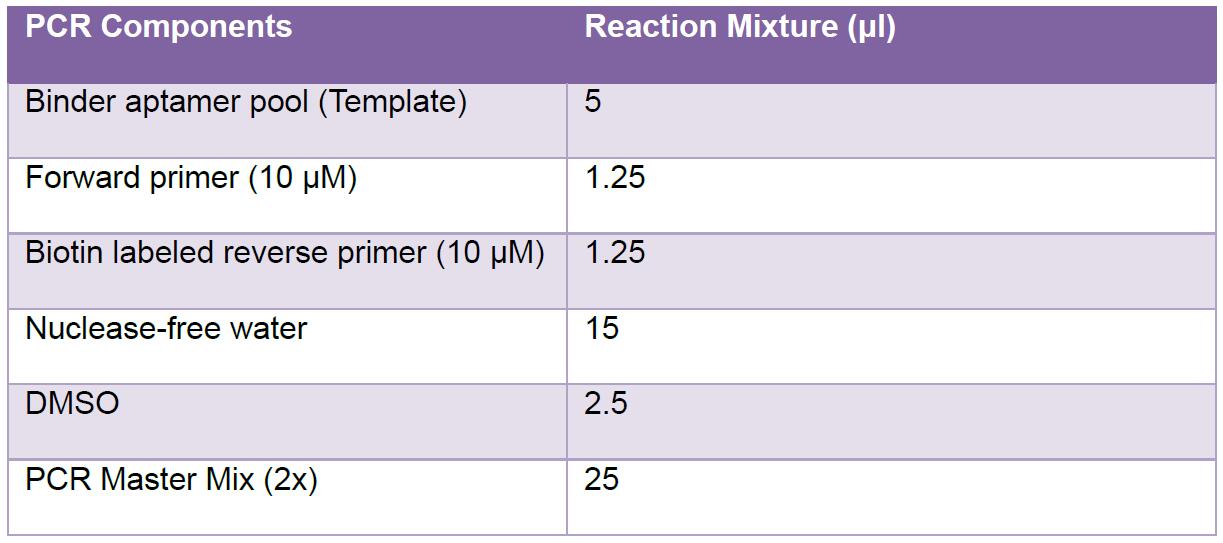
- Run the reaction on a thermocycler preheated at 95 °C using the following program:
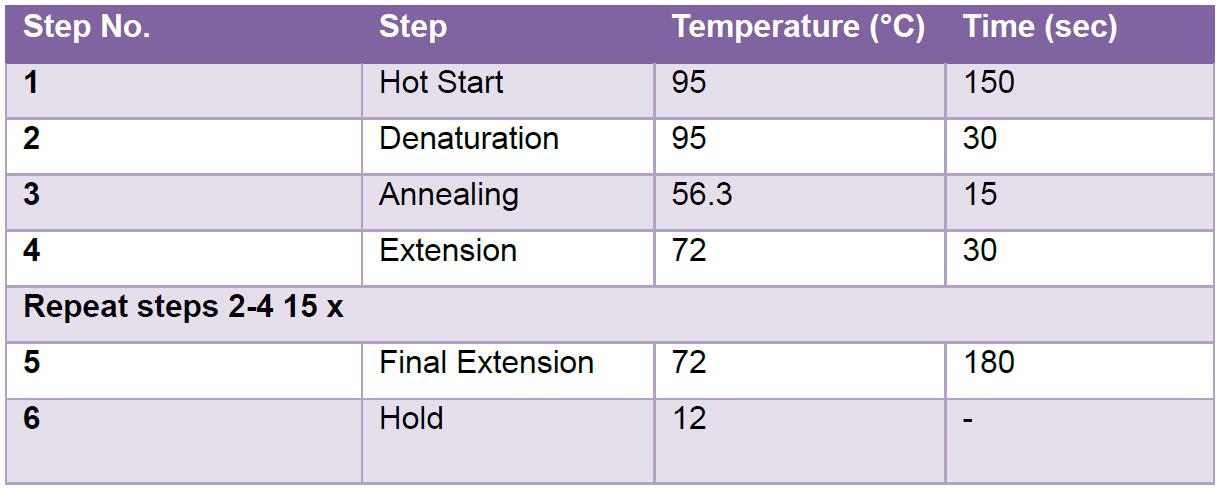
- Heat the PCR product at 95 °C for 10 min.
- Transfer the denatured PCR product to ice immediately and add 5 μl of Streptavidin-Gold bio-probe.
Note: The DNA denaturation (see Step C2 of Day 0) can be repeated by reheating the DNA sample if user fails to transfer the materials to ice-bath. If heating on a heat-block with cooling assembly, use of cooling assembly to chill the samples should be avoided. - Incubate the mixture at 4 °C for 30 min, allowing the streptavidin on the gold nanoparticles to interact with the biotin on the antisense strand.
- Transfer the contents to a 1.5 ml MCT chilled at 4 °C and centrifuge at 14,000 x g for 30 min at 4 °C to separate the sense and antisense aptamer pool strands.
- Harvest the supernatant containing the ssDNA and discard the pellet.
Note: The pellet will be red and the supernatant should be clear. - Quantify the supernatant on NanoVue spectrophotometer to estimate the amount of ssDNA present.
- Store the amplified product at -20 °C till further use.
Days 2-4
SELEX ITERATIVE ROUNDS: Binder exposure
- Dilute the amplified ssDNA binder fraction from Day 2 with binding buffer to a final amount of 1 x 1014 molecules/100 μl (4.44 µg ssDNA/100 µl binding buffer).
- Add to two cell-coated microtiter wells and incubate the microtiter plate for 1 h at 25 °C, similar to the initial round.
- Repeat the steps for screening, amplification and partitioning as previously mentioned in earlier text.
- Repeat the same procedure (the above steps 1-3) till Round 6 by doubling the number of cell-coated wells with each round. (Round 5 will be performed using 16 wells.) Quantify the DNA obtained in each round and compare the increase in DNA concentration. A typical SELEX progression is shown in Figure 2. An increase in binder fractions can be monitored by measuring the concentration of ssDNA aptamer binder pool screened at each round by taking absorbance using a spectrophotometer at λ = 260 nm.
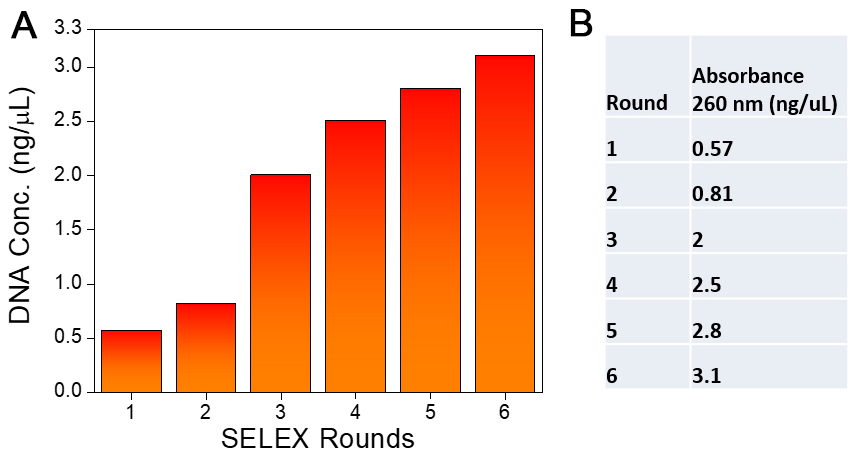
Figure 2. SELEX progression. A. DNA conc. estimation at each SELEX round. B. Table showing assessment of DNA quantity by Nanodrop measurements.
Notes
- This protocol has been optimized for E. coli cells and can be further explored for other cells. It must be noted that the growth media, temperature, duration and shaking conditions should be optimized prior to implementation. The current conditions have been optimized as per the author’s work. For more elaborate understanding of growth curve analysis, users can refer to Wang et al. (2015).
- The viable culture stored at 4 °C can be used for coating the wells within a week. If more time is taken during the SELEX process, new viable culture needs to be generated.
- The pKa of PBA is 8.9 and its binding with glycans is pH dependent. Harsh environmental conditions may affect the binding of the target and PBA.
- To prevent overcrowding and stacking of cells, 1 x 103 cells per well have been optimized with respect to a 96 well plate format. The required cell number enough to cover the well can be estimated by the number of cells (Surface Area = 4πr2) that can cover the internal surface area of the cylindrical well (Internal Surface Area = 2πrh + πr2).
- The length of random region the naïve aptamer library and primers has been found optimal for the target of interest. The forward primer used for amplification can be labeled with fluorophore of choice.
- All the buffers should be prepared in sterile filtered water.
- All centrifugation steps are to be performed at 25 °C unless specified otherwise.
- Number of DNA sequences are calculated for a 81 bases long strand with an average molecular weight of 26,730 g/mol (MWav of single base is 330). The same can be calculated for other sequence lengths by using the multiplication factor.
- With each round the number of binders towards the target molecule increase. If the number of target moieties is not increased, there is a loss of DNA binder population due to the lack of available target sites. To avoid this, the number of target sites is increased following each step.
Recipes
- LB growth media
10 g tryptone
10 g NaCl
5 g yeast extract
Prepare for 1 L
Autoclave at 121.5 °C, 15 psi for 15 min
Cool to 37 °C before inoculation - Carbonate buffer
5.7 mM NaHCO3
4.3 mM Na2HCO3
Prepare for 500 ml volume and adjust pH to 9.8 - Tris-HCl binding buffer
10 mM Tris-HCl
150 mM NaCl
5 mM MgCl2
Prepare for 500 ml volume and adjust pH to 7.6 - Glycine-HCl elution buffer
10 mM glycine
2 mM HCl
Prepare for 100 ml volume and adjust pH to 3.0 - Tris Neutralisation solution
100 mM Tris base
Prepare for 100 ml volume and adjust pH to 11.0 - PBA coating solution
1 mg/ml in carbonate buffer - Phosphate buffered saline
109.9 mg Na2HPO4
27.1 mg NaH2PO4
900 mg NaCl
Prepare for 100 ml volume and adjust pH to 7.4
Acknowledgments
The authors acknowledge DST Nanomission Project (No. SR/NM/NS-1510/2014-G) for financial funding and INST-PhD and CSIR-JRF for fellowships. The protocol described above is adapted from the authors’ work, Kaur et al. (2017).
Competing interests
The authors have no conflict of interest to declare.
References
- Ellington, A. D. and Szostak, J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287): 818-822.
- Kaur, H., Shorie, M., Sharma, M., Ganguli, A. K. and Sabherwal, P. (2017). Bridged rebar graphene functionalized aptasensor for pathogenic E. coli O78:K80:H11 detection. Biosens Bioelectron 98: 486-493.
- Kim, Y. S., Song, M. Y., Jurng, J. and Kim, B. C. (2013). Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal Biochem 436(1): 22-28.
- Priyanka, Shorie, M., Bhalla, V., Pathania, P. and Suri, C. R. (2014). Nanobioprobe mediated DNA aptamers for explosive detection. Chem Commun (Camb) 50(9): 1080-1082.
- Safeh, K., Shangguan, D., Xiong, X., O'Donoghue, M. B. and Tan, W. (2010). Development of DNA aptamers using Cell-SELEX. Nat Protoc 5:1169-1185.
- Tuerk, C. and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968): 505-510.
- Wang, L., Fan, D., Chen, W. and Terentjev, E. M. (2015). Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci Rep 5: 15159.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shorie, M. and Kaur, H. (2018). Microtitre Plate Based Cell-SELEX Method. Bio-protocol 8(20): e3051. DOI: 10.21769/BioProtoc.3051.
Category
Molecular Biology > DNA > DNA-protein interaction
Microbiology > Microbial proteomics > Membrane proteins
Biophysics > Bioengineering > Artificial receptors
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









