- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Examining Autophagy in Plant by Transmission Electron Microscopy (TEM)
Published: Vol 8, Iss 20, Oct 20, 2018 DOI: 10.21769/BioProtoc.3047 Views: 8213
Reviewed by: Zhibing LaiXiaohong ZhuangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Seed Collection in Temperate Trees—Clean, Fast, and Effective Extraction of Populus Seeds for Laboratory Use and Long-term Storage
Naima Bhutta [...] Katharina Bräutigam
Feb 5, 2024 2036 Views

Fast and High-Resolution Imaging of Pollinated Stigmatic Cells by Tabletop Scanning Electron Microscopy
Lucie Riglet and Isabelle Fobis-Loisy
Nov 20, 2024 1785 Views

Cryo-SEM Investigation of Chlorella Using Filter Paper as Substrate
Peng Wan [...] Jinghan Wang
Dec 20, 2024 1448 Views
Abstract
In plants, macroautophagy, here referred as autophagy, is a degradation pathway during which the double-membrane structure named autophagosome engulfs the cargo and then fuses with vacuole for material recycling.
To investigate the process of autophagy, transmission electron microscopy (TEM) was used to monitor the ultrastructure of autophagic structures and identify the cargo during this process due to its high resolution. Compared to other autophagy examination methods including biochemical assays and confocal microscopy, TEM is the only method that indicates the morphology of autophagic structures in nanoscale, which is considered to be one of the best ways to illustrate the morphology of autophagic intermediates and the substrate of autophagy. Here, we describe the autophagy examination assay using TEM in Nicotiana benthamiana leaf cells.
Background
Autophagy is a highly conserved macromolecular degradation pathway in eukaryotes (Dikic, 2017). In plants, autophagy is induced by several stress conditions including starvation, oxidative stress, salt stress and senescence (Doelling et al., 2002; Hanaoka et al., 2002; Liu et al., 2005; Bassham, 2007; Liu and Bassham, 2009; Luo et al., 2017). During autophagy, double-membrane vesicles named autophagosomes form in cytoplasm and transport into the central vacuole, where the outer membrane of autophagosomes fuses with vacuolar membrane. Then the single-membrane structure termed as autophagic body enters into the vacuole lumen and ultimately gets degraded (Ohsumi, 2001; Liu and Bassham, 2012).
Till now, numerous methods for examining autophagy in plants have been established. The frequently used assays are confocal microscopy, electron microscopy and biochemical methods. As for confocal microscopy detection, autophagy marker including ATG8, ATG5 and SH3P2 fused with fluorescent proteins were used to label autophagy related structures (Zhuang et al., 2013; Le Bars et al., 2014; Zhuang and Jiang, 2014; Kliosnky et al., 2016). In addition, fluorescentacidotropic dye such as monodansylcadaverine (MDC) was also used to label autophagic structures in plant cells. Biochemical methods to measure autophagic flux is to detect the ratio of ATG8 and ATG8-PE, or the ratio of GFP-ATG8 and GFP. Nevertheless, TEM is dramatically outstanding among these methods for its high resolution thus provides more legible information of autophagic structure as well as its cargo. Thus, both qualitative and quantitive analysis of autophagy could be performed using TEM observation.
Under TEM observation, autophagosome is distinctly visible as two membrane bilayers which are separated by an electron-translucent aperture (Kliosnky et al., 2016). Meanwhile, it contains cargos for degradation. Generally, during nonselective autophagy, the size of autophagic structures is 0.5-1.5 μm. As for selective autophagy, the size of autophagic structures relies on the specific substrates. Here, we describe the protocol for examining autophagy activity by TEM in Cytoplastic Glyceraldehyde-3-Phosphate (GAPC) silenced plant cells.
Materials and Reagents
- 0.1-10 μl pipette tips (Thermo Fisher Scientific, QSP, catalog number: 104-Q )
- 1-200 μl pipette tips (Thermo Fisher Scientific, QSP, catalog number: 110-B-Q )
- 1-1,000 μl pipette tips (Corning, Axygen®, catalog number: T-1000-B )
- 1.5 ml microcentrifuge tubes
- Toothpick (Yin Sha, catalog number: 918 )
- Grid (Emcn, catalog number: AZH75HH )
- 40 holes flat embedding mold (Emcn, catalog number: DZ10590-40 )
- 4 weeks-old Nicotiana benthamiana plants
- Agrobacterium strain GV3010
- E-64d (Sigma-Aldrich, catalog number: E8640-1MG )
- Paraformaldehyde (Electron Microscopy Sciences, catalog number: 157-8 )
- Glutaraldehyde (Structure Probe, SPI-CHEM, catalog number: 02607-BA )
- Sodium dihydrogen phosphate dehydrate (NaH2PO4•2H2O) (Sigma-Aldrich, catalog number: 1.06342.1000 )
- Disodium hydrogen phosphate dodecahydrate (Na2HPO4•12H2O) (Sigma-Aldrich, catalog number: 04273 )
- Potassium hexacyanoferrate (K3[Fe(CN)6]) (Sigma-Aldrich, catalog number: 702587 )
- OsO4 (Ted Pella, catalog number: 18451 )
- ddH2O
- Uranyl acetate (DEUTSCHL AND LUXEMBURGPRODUZIER TSPE ZIA LCHEMIKALIEN PROD UKTION)
- Lead acetate (Structure Probe, SPI-CHEM, 1161108)
- Ethanol (Sinopharm Chemical Reagent, catalog number: 10009218 )
- Epoxypropane (Sinopharm Chemical Reagent, catalog number: 80059118 )
- 1% (w/v) uranyl acetate in ddH2O and 2% (w/v) uranyl acetate in ddH2O
- DDSA (Structure Probe, SPI-CHEM, catalog number: 02827-AF )
- SPI-PONTM812 (Structure Probe, SPI-CHEM, catalog number: 02659-AB )
- NMA (Structure Probe, SPI-CHEM, catalog number: 02828-AF )
- DMP-30 (Structure Probe, SPI-CHEM, catalog number: 02823-DA )
- HCl (Sinopharm Chemical Reagent, catalog number: 10011008 )
- NaOH (Sinopharm Chemical Reagent, catalog number: 10019762 )
- Trizol reagent (Tiangen)
- RNase-free DNase I (Sigma-Aldrich)
- Oligo(dT)
- TRANScript moloney murine leukemia virus reverse transcriptase (Tiangen)
- Power SYBRGreen PCR master mix (Applied Biosystems)
- Paraformaldehyde-glutaraldehyde Fixative Solution (2%/2.5%), pH 7.2 (see Recipes)
- 0.1 M phosphate buffer (PB), pH 7.2 (see Recipes)
- OsO4-hexacyanoferrate fixative solution (see Recipes)
- 1% (w/v) uranyl acetate (see Recipes)
- 2% (w/v) uranyl acetate (see Recipes)
- 0.2% lead acetate (see Recipes)
- Epon 812 (see Recipes)
- 20 μM E-64d (see Recipes)
Equipment
- Water Purification System (Merck, Milli-Q®, model: Advantage A10 )
- Art knife (FLYING EAGLE, catalog number: 74-S )
- Pincette (Beijing XXBR Technology, catalog number: T5889 )
- Diamond knife (DiATOME, catalog number: DU4535 )
- Eppendorf Research® plus Pipette 0.5-10 μl
- Eppendorf Research® plus Pipette 10-200 μl
- Eppendorf Research® plus Pipette 100-1,000 μl
- Diaphragm vacuum pump (Tianjin Jinteng Experiment Equipment, catalog number: GM-0.50II )
- Shaking incubator (Miulab, catalog number: ES-60 )
- Electronic balance (Sartorius AG)
- Electro-heating standing-temperature cultivator (Tianjin Taisite Instrument, model: DH4000BII )
- Electrothermal constant-temperature dry box (Tianjin Taisite Instrument, model: 202-0AB )
- Ultramicrotome (Leica Microsystems, model: EM UC6 )
- Eyelash pen
- Dyeing machine (Leica Microsystems, model: EM AC20 )
- Electron microscope (Hitachi High-Technologies, model: H-7650 )
- Bio-Rad CFX96 real-time PCR detection system
Procedure
- Nicotiana bentiamiana plants are grown for 4 weeks in growth rooms at 25 °C under a 16-h light/8-h dark cycle.
- Agrobacterium strain GV3010 containing pTRV1 and pTRV2 or its derivative GAPCs or GAPCs/ATG3 co-silencing construct are mixed at the proportion of 1:1 and infiltrated into the top two leaves of the plants to silence GAPCs and GAPCs/ATG3 in plants, respectively.
- When target genes are silenced successfully at 16-18 days post infiltration (dpi), infiltrate 20 μΜ E-64d into the top two new leaves where the genes are silenced successfully.
- After treatment with E-64d for 8-12 h, cut the treated leaves into small pieces (1 mm x 1 mm). Fix the small pieces of leaves in 300 μl paraformaldehyde-glutaraldehyde fixative solution (2%/2.5%, pH 7.2) in a 1.5 ml tube. Vacuumize them using the diaphragm vacuum pump at 0.1 MPa for 10-30 min until the pieces sink to the bottom of the tube. Poke the pieces with toothpicks to help them sink to the bottom of the tube. Fix these pieces with paraformaldehyde-glutaraldehyde fixative solution (2 %/2.5%, pH 7.2) on ice for 1 h.
- Wash the pieces with 0.1 M PBS buffer in a shaker for 3 times, 15 min each time (Steps 5 to 10 should be achieved in a shaker).
- Fix the samples with OsO4-hexacyanoferrate fixative solution (1%/1.5%) on ice for 1 h.
- Wash the pieces with 0.1 M PBS buffer at room temperature (RT) for 3 times, 15 min each time.
- Wash the pieces with ddH2O at RT for 3 times, 10 min each time.
- Stain the samples with 1% uranyl acetatein the dark at RT for 1 h.
- Wash the pieces with ddH2O at RT for 3 times, 15 min each time.
- Dehydrate the samples in a graded series of ethanol of 30%, 50%, 70%, 80%, 90% and 95% (v/v), 10 min each time.
- Dehydrate in 100% ethanol for 3 times, 10 min each time.
- Dehydrate the samples with 100% epoxypropane for 2 times, 10 min each time.
- Incubate the samples with epoxypropane: epon 812 at the volume proportion of 2:1 in a shaker for 30 min at RT.
- Incubate the samples with epoxypropane: epon 812 at the volume proportion of 2:1 in a shaker for 30 min at RT.
- Incubate the samples with epon 812 in a shaker overnight.
- Refresh the epon 812 for 3 times, 3 h each time.
- Embed the samples in 40 holes flat embedding mold with epon 812.
- Permeate the samples with epon 812 at 37 °C for 4 h.
- Aggregate resin at 60 °C, 24-48 h.
- Cut the embedding block with art knife until the sample could be saw. Continue to cut the sample with glass knife until the surface is smooth.
- Cut the sample surface in trapezoidal with an art knife.
- Cut the embedding block into 70-nm ultrathin sections on ultramicrotome with a diamond knife.
- Collect them with eyelash pen on Formvar-coated grids.
- Stain the sections with 2% uranyl acetate for 30 min and 0.2% lead acetate for 5 min using a Dyeing machine.
- Examine the sections under an electron microscope.
- Observe the autophagic structures in mesophyll cells, record the number of autophagic structures and the number of cells.
Data analysis
As shown in Figure 1, a lot of ultrastructure of single-membrane autophagic bodies (arrows) are observed in the vacuole of mesophyll cells of GAPCs-RNAi leaves, whereas few number of autophagic bodies are observed in control or GAPCs/ATG3-RNAi leaves. Virus-induced gene silencing (VIGS) is used here for silencing of GAPCs and ATG3. In Figure 1B, the canonical double-membrane autophagosomes (arrowhead) are observed in the cytoplasm of GAPCs-silenced plants. The black arrowheads show the autophagic bodies inside the vacuole. Figure 1C showed the relative autophagic activity of three samples. Autophagic activity is calculated through dividing the number of totalautophagic structures (both autophagic bodies and autophagosomes) by the number of mesophyll cells. The autophagic activity of TRV control is normalized and set to 1.0. Approximately 20 cells from 2-3 different embedding blocks are used to quantify autophagic structures in each treatment. The statistical analysis indicates the autophagic activity is significantly increased in GAPCs-silenced plants compared to that of control and GAPCs/ATG3-silenced leaves, indicating that silencing of GAPCs activates autophagy, and the activated autophagy is dependent on ATG3.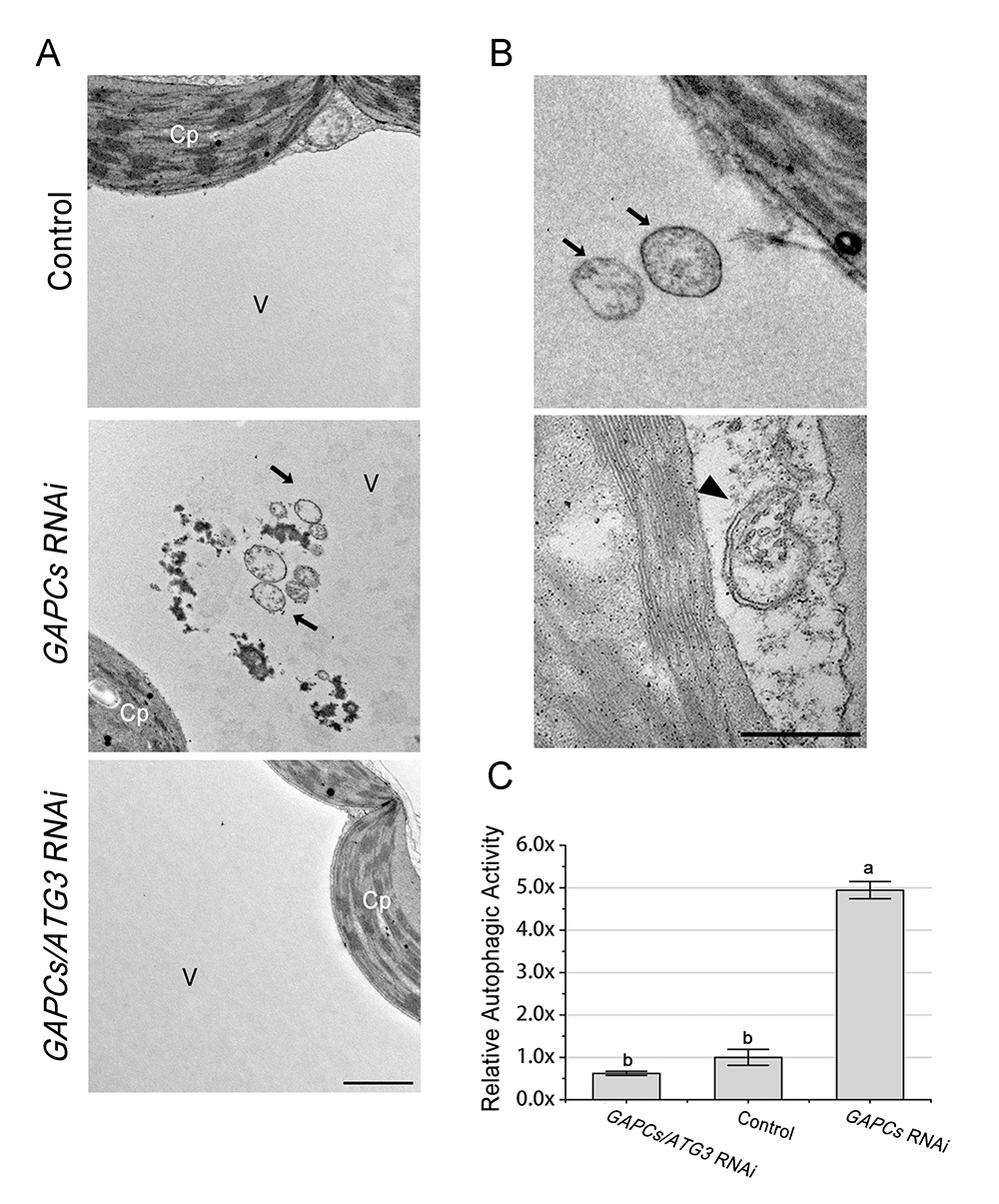
Figure1.Detection of autophagic like structures by TEM in control and VIGS plants. A. Representative TEM images of mesophyll cells from GAPCs-silenced, GAPCs/ATG3-silenced and control leaves. The ultrastructures of autophagic bodies (arrows) are observed in the vacuole of mesophyll cells of GAPCs-silenced plants. Cp, chloroplast; V, vacuole. Scale bar = 1 μm. B. Representative ultrastructure of autophagosomes (arrows) and autophagic bodies (arrowhead) observed in the cytoplasm of mesophyll cells. Scale bar = 500 nm. C. Relative autophagic activity in GAPCs-silenced and GAPCs/ATG3-silenced plants. Autophagic activity in TRV control plants is set to 1.0. Values represent means ± SE from 3 independent experiments. Different letters indicate significant differences (ANOVA, P < 0.05).
Notes
- As plant autophagy is a rapid process, it is helpful to inhibit the autophagy process to allow the observation of transient intermediates (Yoshimoto, 2012). E-64d is a cysteine protease inhibitor which can prevent the autophagic vacuolar degradation thus increase the number of autophagic structures (Liu et al., 2005; Xu et al., 2017). Infiltration of E-64d is optional. It depends on the purpose of the experiment.
- During the preparation of TEM samples, the air humidity should be controlled at a low level (< 40%).
- Samples should be put into the paraformaldehyde-glutaraldehyde fixative solution quickly.
- After application of the paraformaldehyde-glutaraldehyde fixative solution, OsO4-hexacyanoferrate fixative solution and uranyl acetate solution, the samples should be washed thoroughly.
- Alcohol dehydration and resin permeation of samples should be thorough.
- It's crucial to identify autophagic structures properly apart from other double-membrane organelles and vesicles in cytoplasm. Pay attention to the membrane construction, size and the containing substances of the structures. In Nicotiana benthamiana, autophagosomes and autophagic bodies are 0.5-1.5 μm structures. Autophagosomes are double-membrane structures, while autophagosomes fuse with vacuole thus become single-membrane structures termed autophagic bodies. One distinct feature is that, both autophagosomes and autophagic bodies contain substrates for autophagic degradation, such as proteins, organelles, targets lipids, carbohydrates, nucleic acids and invading pathogens (Rabinowitz and White, 2010).
- VIGS is used to silence gene in Nicotiana benthamiana (Liu et al., 2002), and suppress gene expression by infecting Nicotiana benthamiana with a recombinant virus vector carrying host-derived sequence.
- Real-time RT PCR was used to confirm VIGS efficiency of the target genes.
- Extract total RNAs using Trizol reagent (Tiangen) from the leaf samples. Treat the samples with RNase-free DNase I (Sigma-Aldrich) to remove potential DNA contamination. Reverse transcribe RNA using oligo(dT) primer and TRANScript moloney murine leukemia virus reverse transcriptase following the manufacturer’s manual (Tiangen).
- Gene expression analysis. Perform real-time PCR on a Bio-Rad CFX96 real-time PCR detection system using Power SYBR Green PCR master mix (Applied Biosystems). Set real-time PCR cycles to 39. Use actin gene as the internal control (data not shown).
Recipes
Note: All the solutions are prepared with ddH2O unless otherwise stated.
- Paraformaldehyde-glutaraldehyde Fixative Solution
0.1 M phosphate bufferComponents Concentration Paraformaldehyde 2% Glutaraldehyde 2.5%
Adjust pH to 7.2 (with HCl) - 0.1 M phosphate buffer (PB), pH 7.2
Adjust pH to 7.2 with NaOHComponents Concentration NaH2PO4•2H2O 0.028 M Na2HPO4•12H2O 0.072 M - OsO4-hexacyanoferrate fixative solution
Components Concentration OsO4 1% (w/v) Potassium hexacyanoferrate 1.5% (w/v) - 1% (w/v) uranyl acetate
Components Concentration Uranyl acetate 1% - 2% (w/v) uranyl acetate
Components Concentration Uranyl acetate 2% - 0.2% lead acetate
Components Concentration Lead acetate 0.2% NaOH 4% - Epon 812 (100 ml)
Components Content DDSA 16 ml SPI-PONTM 812 50 ml NMA 35.6 ml DMP-30 2 ml - 20 μM E-64d
- E-64d was diluted in ethanol to a 10 mM stock solution and stored at -20 °C
- Dilute10 mM E-64d in ddH2O to the final concentration of 20 μM
Acknowledgments
This protocol was adapted from SJ Han et al.’s work (Han et al., 2015). This work was supported by the National Transgenic Program of China (2016ZX08009-003-001).
Competing interests
The authors declare that there are no conflicts of interest.
References
- Bassham, D. C. (2007). Plant autophagy--more than a starvation response. Curr Opin Plant Biol 10(6): 587-593.
- Dikic, I. (2017). Proteasomal and autophagic degradation systems. Annu Rev Biochem 86: 193-224.
- Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R. and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277(36): 33105-33114.
- Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., Tabata, S. and Ohsumi, Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129(3): 1181-1193.
- Han, S., Wang, Y., Zheng, X., Jia, Q., Zhao, J., Bai, F., Hong, Y. and Liu, Y. (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27(4): 1316-1331.
- Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A. …et al. (2016). Guidelinesfor the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12(1):1-222.
- Le Bars, R., Marion, J., Le Borgne, R., Satiat-Jeunemaitre, B. and Bianchi, M. W. (2014). ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun 5: 4121.
- Liu, Y. and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215-237.
- Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B. and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121(4): 567-577.
- Liu, Y., Schiff, M., Marathe, R. and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30(4): 415-429.
- Liu, Y., Xiong, Y. and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5(7): 954-963.
- Luo, L., Zhang, P., Zhu, R., Fu, J., Su, J., Zheng, J., Wang, Z., Wang, D. and Gong, Q. (2017). Autophagy is rapidly inducedby salt stress and is requiredfor salt tolerance in Arabidopsis. Front Plant Sci 8: 1459.
- Ohsumi, Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2(3): 211-216.
- Rabinowitz, J. D. and White, E. (2010). Autophagy and metabolism. Science 330(6009): 1344-1348.
- Xu, G., Wang, S., Han, S., Xie, K., Wang, Y., Li, J. and Liu, Y. (2017). Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13(7): 1161-1175.
- Yoshimoto, K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53(8): 1355-1365.
- Zhuang, X. and Jiang, L. (2014). Autophagosome biogenesis in plants: roles of SH3P2. Autophagy 10(4): 704-705.
- Zhuang, X., Wang, H., Lam, S. K., Gao, C., Wang, X., Cai, Y. and Jiang, L. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25(11): 4596-4615.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zheng, X., Zhao, C. and Liu, Y. (2018). Examining Autophagy in Plant by Transmission Electron Microscopy (TEM). Bio-protocol 8(20): e3047. DOI: 10.21769/BioProtoc.3047.
Category
Plant Science > Plant physiology > Metabolism
Cell Biology > Cell imaging > Electron microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










