- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Soluble Recombinant Human Tau Protein from Bacteria Using Double-tag Affinity Purification
Published: Vol 8, Iss 22, Nov 20, 2018 DOI: 10.21769/BioProtoc.3043 Views: 9174
Reviewed by: Laia ArmengotAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1814 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1229 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 41 Views
Abstract
Dysfunction of the microtubule-associated protein Tau (encoded by the MAPT gene) has been implicated in more than twenty neurodegenerative diseases, including Alzheimer’s. As such, the physiological and disease-relevant functions of Tau have garnered great interest in the research community. One barrier hampering investigations into the functions of Tau and the generation of pharmacological agents targeting Tau has been the difficulty of obtaining soluble Tau protein in purified form. Here, we describe a protocol that uses dual affinity tag purification to selectively purify soluble recombinant Tau protein from bacteria that is functionally active for downstream applications including immunization, microtubule binding assays, and protein-protein interaction studies.
Keywords: TauBackground
Tau is traditionally defined as a microtubule binding protein; however, in human diseases Tau can dissociate from axonal microtubules and mislocalize to other neuronal compartments including the soma, dendrites, and synapses, where interactions with non-microtubule proteins and structures drive neuronal dysfunction (Iqbal et al., 2016; Wang and Mandelkow, 2016; Zhou et al., 2017; McInnes et al., 2018). Although Tau aggregates in the form of neurofibrillary tangles are commonly found in post-mortem diseased brain tissue, studies suggest that soluble Tau, not aggregated Tau, is largely responsible for neuronal dysfunction (Crimins et al., 2012; Polydoro et al., 2014; Koss et al., 2016). As such, investigating the soluble functions of Tau in disease, such as identifying protein-protein interaction partners, is therefore of critical importance to target Tau dysfunction.
Purifying soluble Tau protein has been challenging since many ectopically expressed recombinant proteins aggregate into insoluble inclusion bodies in bacteria. Tau furthermore contains aggregation-prone motifs, making purifying recombinant Tau even more difficult. We have therefore developed a protocol that overcomes these obstacles by optimizing two key aspects of this process: first, we express Tau as a fusion protein with an N-terminal Glutathione S-transferase (GST) tag to enhance solubility of the protein, and utilize a mild induction paradigm to minimize the formation of insoluble Tau aggregates. We chose GST over other fusion proteins because it is relatively small, robustly enhances protein solubility and stability, can be easily cleaved, and is relatively inexpensive to purify. Second, our purification protocol utilizes dual affinity tag purification of both an N-terminal GST tag and a C-terminal 8xHis tag (Figure 1); because this purification protocol requires both N- and C-terminal tags to be available to bind affinity resin, the protocol therefore specifically enriches for soluble, non-aggregated protein (in the event of aggregation, at least one of these epitopes is hidden within the aggregate). Following the first purification step against the N-terminal GST tag, the GST fusion protein is cleaved using PreScission protease, resulting in a final product of purified Tau protein that contains only an additional eight histidine residues (8xHis tag) at its C-terminal domain that can also be used for downstream applications including pull-down experiments or detection using anti-His antibodies. This method is versatile and can be easily modified to purify different isoforms of Tau, Tau carrying point mutations, or Tau truncations by altering the pGEX_GST-Tau0N4R-8xHis plasmid using site-directed mutagenesis or traditional cloning methods. We have successfully utilized this protocol to produce various isoforms of Tau, including with point mutations or domain truncations, for use in in vitro binding assays with synaptic vesicles (Zhou et al., 2017) and as bait in protein-protein interaction studies (McInnes et al., 2018).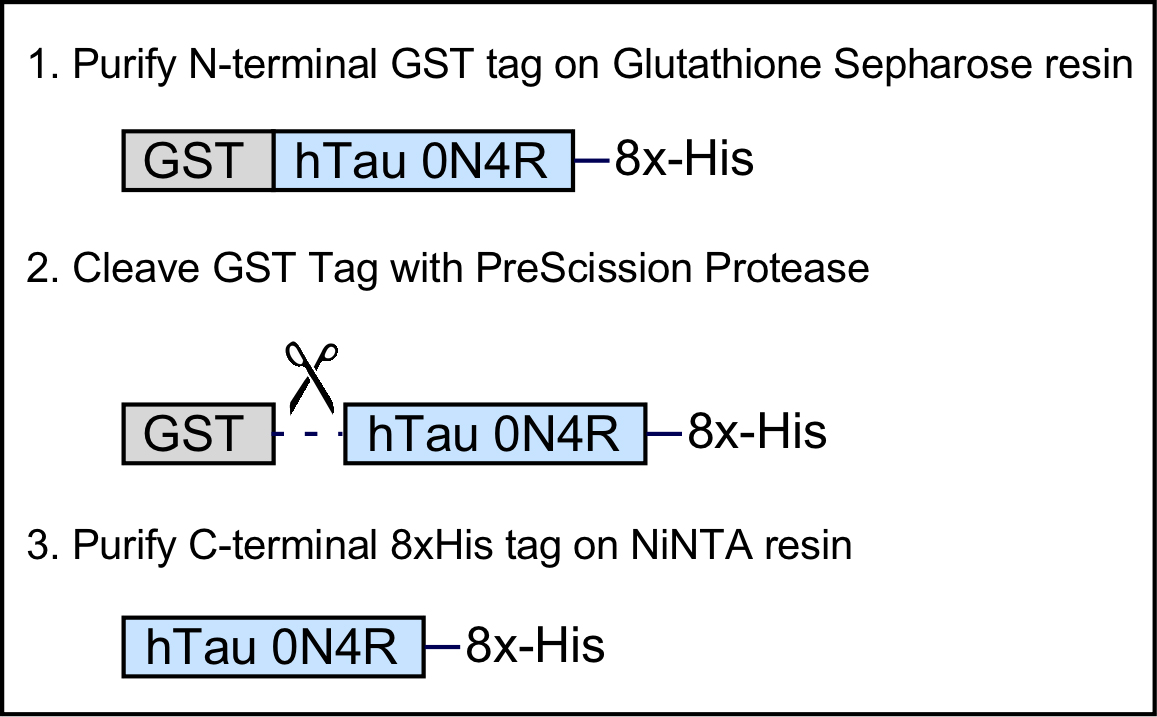
Figure 1. Schematic overview of the recombinant GST-Tau-8xHis fusion protein and purification steps
Materials and Reagents
- 1 ml micropipette tips
- Microcentrifuge tubes
- Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-10 membrane (Merck, catalog number: UFC501024 )
- Plasmid pGEX_GST-Tau0N4R-8xHis
Note: This plasmid can be generated by cloning a cDNA encoding human MAPT into a pGEX-6P-1 vector backbone (such as the BamHI/EcoRI restriction sites of Addgene plasmid number 46408). The cDNA should be at downstream of N-terminal GST fusion with PreScission Protease cleavage site. The 8xHis tag can be inserted into the 3’ primer used to amplify the hTau cDNA. In this protocol, we use human MAPT cDNA corresponding to the 0N4R isoform. - RosettaTM bacteria cells (Merck, catalog number: 70953 )
- LB Broth (Sigma-Aldrich, catalog number: L3522 )
- Ampicillin (Sigma-Aldrich, catalog number: A9393 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Anti-His antibody (such as Thermo Fisher Scientific, catalog number: 37-2900 ) or anti-Tau antibody (such as Agilent Technologies, DAKO, catalog number: A0024 )
- Isopropyl β-D-thiogalactoside (IPTG) (Sigma-Aldrich, catalog number: I6758 )
Note: Prepare 100 mM stock in sterile H2O and freeze aliquots at -20 °C. Do not re-freeze after thawing. - Glutathione Sepharose 4B (GE Healthcare, catalog number: 17075601 )
- Complete protease inhibitor tablets, EDTA-free (Roche Diagnostics, catalog number: 11873580001 )
- Phenylmethanesulfonyl fluoride (PMSF), 100 mM solution (Sigma-Aldrich, catalog number: 93482 )
- Benzonase Nuclease, 250 units/μl (Sigma-Aldrich, catalog number: E1014 )
- PreScission Protease, 2,000 units/ml (GE Healthcare, catalog number: 27084301 )
Note: Upon receiving stock, thaw on ice and prepare ~10 μl aliquots and freeze at -20 °C. Do not re-freeze aliquots after thawing. - Ni-NTA ProfinityTM IMAC Resin, Ni-charged (Bio-Rad Laboratories, catalog number: 1560131 )
- Quick StartTM Bradford Protein Assay (Bio-Rad Laboratories, catalog number: 5000201 )
- PageBlueTM Protein Staining Solution (Thermo Fisher Scientific, catalog number: 24620 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Lysozyme (Sigma-Aldrich, catalog number: L6876 )
- Sodium phosphate dibasic anhydrous (Na2HPO4) (Sigma-Aldrich, catalog number: 795410 )
- DL-Dithiothreitol (DTT), 1 M stock solution in H2O (Sigma-Aldrich, catalog number: 646563 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E6758 )
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S8045 )
- Imidazole (Sigma-Aldrich, catalog number: 792527 )
- Bacteria lysis buffer (see Recipes)
- Phosphate buffered saline (PBS) (see Recipes)
- PBS + 250 mM NaCl (see Recipes)
- PreScission protease cleavage buffer (see Recipes)
- Ni-NTA wash buffer (see Recipes)
- Ni-NTA elution buffer (see Recipes)
Equipment
- Pipettes
- Table-top microcentrifuge capable of speeds up to 16,000 x g
- Microplate reader or spectrophotometer capable of absorbance readings at 600 nm for bacterial density measurements and at 595 nm for Bradford protein assay
Procedure
Notes:
- The following procedure is given for purifying Tau protein from a 50 ml culture volume of bacteria, which yields approximately 4 μg pure protein. This protocol can easily be scaled up if more protein is required.
- To guarantee the solubility of Tau, it is essential that the protein is purified fresh and is used for experiment immediately upon finishing purification. Freezing purified Tau protein nucleates aggregation and results in loss of solubility and function.
- Perform all purification procedures at 4 °C and/or on ice, and pre-chill all tubes on ice.
- Bacterial cell induction and harvest
- Transform a suitable bacterial strain for protein production with the pGEX_GST-Tau0N4R-8xHis plasmid (ampicillin-resistant). We recommend using Rosetta bacteria that carry a chloramphenicol-resistant tRNA plasmid to enhance expression of mammalian proteins.
- Starter culture: In the afternoon, inoculate 5 ml LB media containing appropriate antibiotics (100 μg/ml ampicillin for pGEX_GST-Tau0N4R-8xHis plasmid; additionally, 25 µg/ml chloramphenicol if using Rosetta cells, highly recommended) with a single colony of bacteria (or 10 μl from a glycerol stock) and let grow overnight at 37 °C with shaking at 220-250 rpm.
- Main culture: The following morning, dilute the saturated overnight 5 ml culture (OD600 ~2.0) into 45 ml fresh LB medium containing appropriate antibiotics (1:10 dilution, to OD600 ~0.2), and incubate for 1 h at 37 °C with shaking at 220-250 rpm to allow bacteria to recover to exponential growth phase.
- After 1 h, add IPTG to a final concentration of 0.4 mM and let bacteria grow at 37 °C for 2 h.
Optional: Confirm bacteria have re-entered exponential growth phase by measuring the density of the culture at OD600. Bacteria should have recovered from OD600 ~0.2 to at least OD600 ~0.4 before induction. - Two hours following induction, pellet bacteria by centrifugation at 5,000 x g for 15 min at 4 °C. Freeze bacterial cell pellet at -80 °C until use.
Note: Bacterial cell pellets are stable for at least 3 months at -80 °C. We routinely prepare 0.5-2 L batches of bacteria, freeze 50 ml cell culture volume pellets in aliquots, and take single aliquots for purification on experiment days.
- Bacterial cell lysis
- Take a single bacterial cell pellet from a 50 ml culture and thaw on ice. Thoroughly resuspend pellet in 1.5 ml ice-cold bacteria lysis buffer by pipetting up and down using 1 ml micropipette tip that has been cut to make it wide-bore and transfer the lysate to a microcentrifuge tube on ice.
Note: Be sure to freshly prepare lysis buffer (see Recipes). - Incubate lysate for 30 min at 4 °C on a rotating wheel to allow for complete lysis.
- Centrifuge lysate at 16,000 x g for 20 min at 4 °C to pellet insoluble material and debris, and transfer the supernatant (cleared lysate) to a new microcentrifuge tube on ice. Complete lysis will be evident as the lysate becomes light brown in color.
- Take a single bacterial cell pellet from a 50 ml culture and thaw on ice. Thoroughly resuspend pellet in 1.5 ml ice-cold bacteria lysis buffer by pipetting up and down using 1 ml micropipette tip that has been cut to make it wide-bore and transfer the lysate to a microcentrifuge tube on ice.
- Purification against N-terminal GST tag
- Preparation of glutathione sepharose: Using a cut pipette tip, transfer 75 μl of Glutathione Sepharose 4B slurry into a microcentrifuge tube and add 1 ml ice-cold PBS to wash.
Note: Make sure glutathione sepharose slurry is thoroughly mixed by gentle shaking for 30 sec before using. - Pellet sepharose by centrifugation at 3,500 x g for 3 min.
- Discard supernatant. Wash sepharose again with 1 ml ice-cold PBS, centrifuge and discard supernatant.
- Transfer the total volume of the cleared bacterial lysate (approximately 1.5 ml lysate from 50 ml bacterial culture) into the tube containing washed glutathione sepharose beads.
- Incubate lysate with glutathione sepharose for 2 h at 4 °C with rotation to allow GST tag to bind glutathione.
- After binding, pellet sepharose (3,500 x g for 3 min at 4 °C) and wash with 1 ml of PBS + 250 mM NaCl. Pellet again and discard supernatant. Repeat washing 2 more times.
- Preparation of glutathione sepharose: Using a cut pipette tip, transfer 75 μl of Glutathione Sepharose 4B slurry into a microcentrifuge tube and add 1 ml ice-cold PBS to wash.
- Cleavage of the N-terminal GST tag
- Prepare PreScission protease cleavage buffer by adding fresh DTT to a final concentration of 1 mM immediately before use.
- Equilibrate beads (GST-Tau-8xHis bound sepharose) by washing twice each with 1 ml PreScission protease cleavage buffer with DTT (same washing procedure as in Procedure C).
- After the second wash, discard supernatant and resuspend bead slurry in 240 μl PreScission protease cleavage buffer with DTT.
- Transfer the suspension to a smaller tube (e.g., 0.5 ml tube) and add 10 μl (20 U) PreScission protease.
- Incubate the cleavage mixture overnight at 4 °C with rotation. Ensure tube is of appropriate volume to allow for sufficient mixing of the reaction.
Note: The minimum cleavage time recommended by GE Healthcare is 4 h at 4 °C, but we recommend overnight incubation.
- Purification against the C-terminal His tag
- The next morning, pellet the sepharose slurry by centrifugation at 3,500 x g for 5 min at 4 °C. Transfer the supernatant to a new microcentrifuge tube and discard the sepharose bead slurry. This supernatant contains the cleaved Tau-His as well as residual PreScission protease.
- Prepare fresh glutathione sepharose slurry (50 μl), wash twice in cleavage buffer containing DTT, then incubate supernatant from Step E1 wish fresh glutathione sepharose for 1 h at 4 °C with mixing.
Note: This step will remove the residual GST-tagged PreScission protease as well as any residual uncleaved GST-Tau-His protein in the mixture. - After 1 h, pellet the sepharose by centrifugation at 3,500 x g for 5 min at 4 °C, and transfer the supernatant, which contains the free Tau-His protein, to a new microcentrifuge tube on ice.
- Prepare Ni-NTA resin by pipetting 25 μl of Ni-NTA slurry into a microcentrifuge tube and wash twice in ice-cold PreScission protease cleavage buffer to equilibrate the resin.
- Apply the supernatant containing Tau-His protein to the Ni-NTA resin and allow to bind for 45 min at 4 °C with mixing.
- After 45 min, pellet Ni-NTA slurry at 3,500 x g for 3 min at 4 °C and discard supernatant. Wash beads three times with ice-cold 1 ml Ni-NTA wash buffer.
- Elute the bound Tau-His protein by resuspending washed beads in ice-cold 500 μl Ni-NTA elution buffer and incubating for 10 min at 4 °C with rotation.
- Pellet Ni-NTA beads by centrifugation at 10,000 x g for 2 min at 4 °C. Collect the supernatant, being careful not to transfer any sedimented Ni-NTA beads, and transfer to a new microcentrifuge tube on ice.
- Concentrate the 500 μl protein eluate down to a volume of approximately 100 μl by applying the eluate to an Amicon Ultra-0.5 ml Centrifugal Filter Unit (30 kDa molecular weight cutoff) and centrifuging at 14,000 x g for approximately 8 min at 4 °C, or longer until the volume has reached 100 μl. Recover solution by inverting the filter unit into a clean collection tube and centrifuging at 1,000 x g for 2 min at 4 °C.
- Measure protein concentration using the Bradford Protein Assay, or other appropriate protein quantification method. Confirm protein purity by SDS-PAGE and staining with PageBlue colloidal Coomassie reagent.
Note: Expect the protein concentration to be approximately 20-60 μg/ml, which requires putting up to 10 μl of protein sample into a 200 μl Bradford reaction for detection.
Data analysis
- To evaluate the purity of the final product, run 1 μg protein on an SDS-PAGE gel and stain with colloidal Coomassie. We routinely observe greater than 90% protein purity using this protocol (Figure 2).
- The identity of the purified protein product can be confirmed by immunoblotting using an anti-His antibody or anti-Tau antibody. For examples of immunoblots detecting full-length and truncated Tau proteins purified using this protocol see Zhou et al. (2017) and McInnes et al. (2018).
- The aggregation status of purified Tau protein can be assessed by ultracentrifugation of the sample at 100,000 x g for 1 h at 4 °C, followed by analyzing the presence of Tau aggregates in the pellet by SDS-PAGE. Analyzing freshly purified protein using this protocol, we observe minimal Tau protein in the insoluble fraction. In contrast, freezing and thawing the purified protein results in abundant Tau protein in the pellet upon centrifugation at 100,000 x g, indicating aggregation.
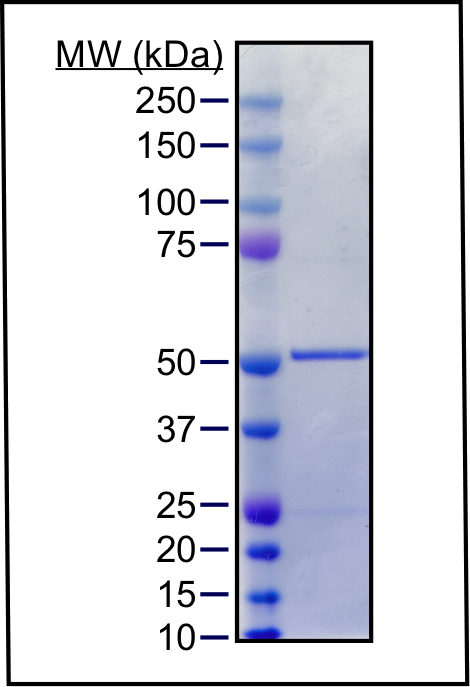
Figure 2. Purity of Tau-His protein. One microgram purified protein was mixed with 1x NuPAGE LDS sample buffer (InvitrogenTM) containing 1% β-mercaptoethanol, heated at 70 °C for 10 min, and separated on a NuPAGE 4-12% Bis-Tris Protein Gel (InvitrogenTM) with MOPS running buffer. The gel was stained with PageBlue colloidal Coomassie solution.
Recipes
- Bacteria lysis buffer
1x PBS (from 10x stock solution)
1% Triton X-100 (from 10% stock solution)
10% glycerol (from 50% stock solution)
1 mg/ml lysozyme (dissolve fresh from powder)
1x Complete Protease Inhibitor cocktail, EDTA-free (Roche Diagnostics)
1 mM PMSF (from 100 mM stock solution)
250 U/ml Benzonase (1:1,000 from enzyme stock)
Note: Add Complete protease inhibitor cocktail, PMSF, lysozyme and Benzonase fresh immediately before use. - Phosphate buffered saline (PBS)
10 mM Na2HPO4
2.7 mM KCl
137 mM NaCl
Adjust pH to 7.4 - PBS + 250 mM NaCl
Dilute a 5 M stock of NaCl into PBS to give a final concentration of 250 mM NaCl - PreScission protease cleavage buffer
20 mM Tris-HCl
50 mM NaCl
0.5 mM EDTA
0.01% Tween-20
1 mM DTT
Adjust pH to 7.0 with NaOH
Notes:- Buffer (without DTT) can be kept at 4 °C for up to 1 month.
- Immediately before use, take an aliquot of buffer and supplement with DTT.
- Ni-NTA wash buffer
50 mM NaH2PO4
300 mM NaCl
20 mM imidazole
pH 8.0
Store at 4 °C - Ni-NTA elution buffer
50 mM NaH2PO4
300 mM NaCl
250 mM imidazole
pH 8.0
Store at 4 °C
Acknowledgments
This work was supported by a European Research Council consolidator grant (646671), the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO), and an academic-industrial collaboration between the VIB-KU Leuven Center for Brain and Disease Research and Janssen Pharmaceutical Companies of Johnson & Johnson.
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Crimins, J. L., Rocher, A. B. and Luebke, J. I. (2012). Electrophysiological changes precede morphological changes to frontal cortical pyramidal neurons in the rTg4510 mouse model of progressive tauopathy. Acta Neuropathol 124(6): 777-795.
- Iqbal, K., Liu, F. and Gong, C. X. (2016). Tau and neurodegenerative disease: the story so far. Nat Rev Neurol 12(1): 15-27.
- Koss, D. J., Jones, G., Cranston, A., Gardner, H., Kanaan, N. M. and Platt, B. (2016). Soluble pre-fibrillar tau and beta-amyloid species emerge in early human Alzheimer's disease and track disease progression and cognitive decline. Acta Neuropathol 132(6): 875-895.
- McInnes, J., Wierda, K., Snellinx, A., Bounti, L., Wang, Y. C., Stancu, I. C., Apostolo, N., Gevaert, K., Dewachter, I., Spires-Jones, T. L., De Strooper, B., De Wit, J., Zhou, L. and Verstreken, P. (2018). Synaptogyrin-3 mediates presynaptic dysfunction induced by Tau. Neuron 97(4): 823-835 e8.
- Polydoro, M., Dzhala, V. I., Pooler, A. M., Nicholls, S. B., McKinney, A. P., Sanchez, L., Pitstick, R., Carlson, G. A., Staley, K. J., Spires-Jones, T. L. and Hyman, B. T. (2014). Soluble pathological tau in the entorhinal cortex leads to presynaptic deficits in an early Alzheimer's disease model. Acta Neuropathol 127(2): 257-270.
- Wang, Y. and Mandelkow, E. (2016). Tau in physiology and pathology. Nat Rev Neurosci 17(1): 5-21.
- Zhou, L., McInnes, J., Wierda, K., Holt, M., Herrmann, A. G., Jackson, R. J., Wang, Y. C., Swerts, J., Beyens, J., Miskiewicz, K., Vilain, S., Dewachter, I., Moechars, D., De Strooper, B., Spires-Jones, T. L., De Wit, J. and Verstreken, P. (2017). Tau association with synaptic vesicles causes presynaptic dysfunction. Nat Commun 8: 15295.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
McInnes, J., Zhou, L. and Verstreken, P. (2018). Purification of Soluble Recombinant Human Tau Protein from Bacteria Using Double-tag Affinity Purification. Bio-protocol 8(22): e3043. DOI: 10.21769/BioProtoc.3043.
Category
Neuroscience > Cellular mechanisms > Protein isolation
Neuroscience > Nervous system disorders > Cellular mechanisms
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












