- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enzymatic Synthesis and Fractionation of Fluorescent PolyU RNAs
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2988 Views: 5230
Reviewed by: Vamseedhar RayaproluKarthik KrishnamurthyOmar Akil

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Versatile Click Chemistry-based Approaches to Illuminate DNA and RNA G-Quadruplexes in Human Cells
Angélique Pipier and David Monchaud
Feb 5, 2025 2534 Views

RNA PolyA Tailing Assay to Qualitatively Analyze Circular RNA Manufacturing
Zih-Shiuan Chuang [...] Chia-Yi Yu
May 20, 2025 1653 Views

APEX2 RNA Proximity Labeling in Mammalian Cell Lines With Low Biotin Permeability
Adrian Beat Tschan [...] Alexa B.R. McIntyre
Jul 5, 2025 2322 Views
Abstract
The physical properties of viral-length polyuridine (PolyU) RNAs, which cannot base-pair and form secondary structures, are compared with those of normal-composition RNAs, composed of comparable numbers of each of A, U, G and C nucleobases. In this protocol, we describe how to synthesize fluorescent polyU RNAs using the enzyme polynucleotide phosphorylase (PNPase) from Uridine diphosphate (UDP) monomers and how to fractionate the polydisperse synthesis mixture using gel electrophoresis, and, after electroelution, how to quantify the amount of polyU recovered with UV-Vis spectrophotometry. Dynamic light scattering was used to determine the hydrodynamic radii of normal-composition RNAs as compared to polyU. It showed that long polyU RNAs behave like linear polymers for which the radii scale with chain length as N1/2, as opposed to normal-composition RNAs that act as compact, branched RNAs for which the radii scale as N1/3.
Keywords: Homopolymer polyU RNABackground
PolyU as a Physical Object: PolyU is an abiological RNA molecule composed of repeated uridine residues, and therefore, it cannot Watson-Crick base-pair and is devoid of RNA secondary structure (Martin and Ames, 1962; Richards et al., 1963). PolyU has very weak base-stacking energy resulting in a lack of helical ordering as well–except below 4 °C (Richards et al., 1963). Due to this lack of structure polyU RNA can be described physically as a random coil (Inners and Felsenfeld, 1970), as opposed to normal RNA molecules that form complicated secondary/tertiary structures, making them more compact physical objects (Yoffe et al., 2008; Fang et al., 2011; Gopal et al., 2012; Garmann et al., 2015).
We therefore expect polyU to be less compact than viral RNA of the same length. Indeed dynamic light scattering (DLS) measurements (see Figure 3) indicate that in RNA assembly buffer (see Recipes) at 25 °C, polyU molecules are about 25% larger in average diameter than branched RNAs of the same length. Additionally, we expect polyU to be more flexible than normal-composition RNAs, as it lacks secondary and tertiary structure, resulting in a more extended, flexible molecule.
Synthesis and Fractionation of Long PolyU RNAs: The synthesis protocol used herein was developed from existing protocols used by Vanzi et al. (2003) and van den Hout et al. (2011). Specifically, polyU RNAs were synthesized from Uridine diphosphate (UDP) monomers using the enzyme polynucleotide phosphorylase (PNPase), which indiscriminately adds nucleotide diphosphates (NDPs) to the 3' end of oligonucleotides, in this case a 20-nt-long polyU seed oligo with a 5' cy5 fluorescent label (Beren et al., 2017). It is important to note that the oligo sequence and length can be changed as desired, and it can be tagged at the 5' end with fluorophores, linkers, or modified bases, enabling the synthesis of polyU with customizable markers. Because of its lack of base-pairing, polyU cannot be visualized in electrophoresis gels by the usual intercalating nucleic acid stains. For this reason, we chose to utilize a fluorophore at the 5' end of the RNA. Additionally, it is possible to make polyA, polyG or polyC using the same PNPase, by simply adding ADP, GDP or CDP, respectively (Grunberg-Manago et al., 1956).
The synthesis reactions produce a polydisperse mixture of fluorescent polyU RNAs ranging in length from 200 to 10,000 nts, with shorter lengths being represented in significantly higher copy number as determined by fluorescence detection in denaturing agarose gel electrophoresis (see Figure 1). After running the RNA mixture alongside a denatured ssRNA ladder (see Figure 1), it was fractionated by electroeluting excised portions of the gel using the ladder as a reference. In fact, we often run many lanes of polyU in parallel to increase the amount of fractionated polyU produced from a given gel, as we usually recover between 30% and 60% of the RNA after gel electroelution.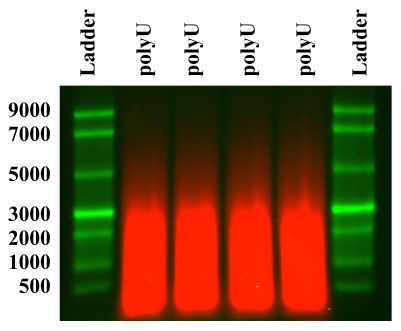
Figure 1. Denaturing electrophoresis gel analysis of unfractionated polyU. Electrophoresis gel analysis of unfractionated polyU (red, cy5 fluorescence) after synthesis using PNPase, compared with a single-stranded (ss) RNA ladder (green, ethidium bromide nucleic acid stain). This gel illustrated the high amount of polydispersity in the polyU generated from the synthesis reaction. The brightest band in the RNA ladder is 3,000-nt-long ssRNA.
We used this process to fractionate polyU into the following length fractions: 500-1,500, 1,500-2,500, 2,500-3,500, 3,500-5,000, 5,000-7,000 and 7,000-9,000 nt. The amount of polyU purified was quantified using UV-Vis absorbance measurements, and the absorbance ratio at 260 nm/280 nm was used as a measure of RNA purity (A260/A280 greater than 2.0). These fractionated polyU samples were then examined on a denaturing agarose gel to demonstrate that single contiguous RNA bands were obtained (see Figure 2).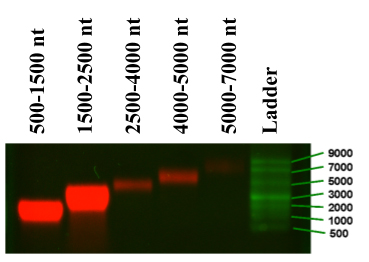
Figure 2. Denaturing electrophoresis gel analysis of fractionated polyU. From right to left: ssRNA ladder, 5,000-7,000, 4,000-5,000, 2,500-4,000, 1,500-2,500 and 500-1,500 nt polyU RNAs. The decrease in band intensity for higher-molecular-weight polyU molecules is due to the fact that an equal mass of polyU was loaded per lane, resulting in many fewer RNAs in the higher-molecular-weight fractions, and subsequently, a decreased fluorescence signal (because of there being only one label per RNA molecule). (Adapted from Beren et al., 2017)
Materials and Reagents
- PolyU synthesis
- 100 kDa Amicon filters (Merck, catalog number: UFC510096 )
- 6-8 kDa MW cutoff dialysis tubing (Repligen, Spectra/Por®, catalog number: 132650 )
- Eppendorf tubes (Eppendorf, catalog number: 022363204 )
Note: We do not utilize RNase-free tubes, but always autoclave the Eppendorf tubes before use. - RNase Inhibitor (Roche Diagnostics, catalog number: 03 335 399 001 , stored at -20 °C)
- Polynucleotide phosphorylase (PNPase) from Synechocystis sp. (Sigma-Aldrich, catalog number: N9914-100UG , stored at -80 °C)
- Uridine diphosphate (UDP, Sigma-Aldrich, catalog number: 94330 , stored at -20 °C)
- 10-25 nt RNA seed oligo (IDT, DNA, stored in TE buffer at -80 °C)
Note: We utilized a 20-nt-long polyU RNA seed oligo for our experiments. - Tris base (Merck, catalog number: 648310-2.5KG ), for making buffer Tris pH 9
- EDTA (Merck, Omnipur®, catalog number: 4050-1KG )
- Magnesium chloride hexahydrate (MgCl2•6H2O) (Merck, Omnipur®, catalog number: 5980-500GM )
- Hydrochloric Acid (Thermo Fisher Scientific, catalog number: FLA144500 )
- Synthesis buffer (stored at 4 °C) (see Recipes)
- TE buffer (stored at 4 °C) (see Recipes)
- PolyU fractionation
- 100 kDa Amicon filters (Merck, catalog number: UFC510096 )
- 6-8 kDa MW cutoff dialysis tubing (Repligen, Spectra/Por®, catalog number: 132650 )
- Eppendorf tubes (Eppendorf, catalog number: 022363204 )
Note: We do not utilize RNase-free tubes, but always autoclave the Eppendorf tubes before use. - Dialysis tubing clips (Repligen, Spectra/Por®, catalog numbers: 132743 and 132735 )
- Sodium bicarbonate (Merck, catalog number: SX0320-1 )
- RNase Inhibitor (Roche Diagnostics, catalog number: 03 335 399 001 , stored at -20 °C)
- Polynucleotide phosphorylase (PNPase) from Synechocystis sp. (Sigma-Aldrich, catalog number: N9914-100UG , stored at -80 °C)
- ssRNA ladder (New England Biolabs, catalog number: N0362S , stored at -80 °C)
- 1% agarose gel (prepared in TAE buffer, agarose is purchased from Merck, Omnipur®, Calbiochem, catalog number: 2125-500GM )
- Formamide (Acros organics, catalog number: 327235000 , stored at 4 °C)
- Gel red (Biotium, catalog number: 41001 )
- Tris base (Merck, catalog number: 648310-2.5KG ), for making buffer Tris pH 8
- Acetic acid (Sigma-Aldrich, catalog number: 695092 )
- EDTA (Merck, Omnipur®, catalog number: 4050-1KG )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S671-10 )
- Potassium chloride (KCl) (Merck, catalog number: 7300-500GM )
- Magnesium chloride hexahydrate (MgCl2•6H2O) (Merck, Omnipur®, catalog number: 5980-500GM )
- Hydrochloric Acid (Thermo Fisher Scientific, catalog number: FLA144500 )
- TAE buffer (stored at 4 °C) (see Recipes)
- RNA assembly buffer (see Recipes)
Equipment
Note: Everything is sterilized before use, either by autoclaving or rinsing with 70% ethanol.
- Scalpel (Surgical Design, model: SCALPEL#15 )
- 70% Ethanol (We dilute 100% ethanol with ddH2O) (Decon Labs, catalog number: V1001 )
- Micropipette volumes used range from 1-1,000 μl (Mettler-Toledo, Rainin, model: Pipet-Lite XLS LTS )
- Micropipette tips: volumes used range from 1-1,000 μl (filtered, sterile aerosol tips with high recovery for Rainin LTS Pipettemen; VWR, catalog numbers: 83009-688 , 82003-196 , 82003-198 )
- Autoclave
- Heat block (set to 70 °C) (Fisher Scientific, catalog number: 11-718-2Q )
- Ice or a cold block for RNA work (VWR, catalog number: 414004-285 )
- 37 °C incubator (Precision, catalog number: 51221089 )
- Centrifuge that can run to 5,000 x g (Eppendorf, model: 5804 R )
- Gel electrophoresis equipment
Gel tank/box combo (Fisher Scientific, catalog number: FB-SB-710 )
Gel combs (Thermo Fisher Scientific, catalog numbers: B1A-10 and B1A-6 )
Power supply (Fisher Scientific, catalog number: FB1000Q ) - Laminar flow hood, for RNA work
- UV lamp, for imaging the ssRNA ladder in the gel (Spectronics, model: ENF-260C )
- Nanodrop, UV-Vis spectrophotometer for measuring RNA concentrations
- Malvern particle sizer Nano-ZS
- Milli-Q UV Plus (Merck, catalog number: ZD60115UV )
Procedure
Note: Samples are kept on ice unless otherwise specified, and all work is done in a sterile, laminar flow hood to prevent contamination of the RNA. All buffers, Eppendorf tubes, and glassware are autoclaved before use.
- Preparing dialysis tubing
- Prepare a solution of 2% sodium bicarbonate, 1 mM EDTA is in a glass beaker.
- Cut the Dialysis tubing into 4-in segments and boil in the solution of 2% sodium bicarbonate, 1 mM EDTA for 10 min. Rinse the tubing thoroughly with ddH2O, and then boil for 10 min in ddH2O.
- Cover the tubing with ddH2O once again. Autoclave, and store at 4 °C.
- PolyU synthesis
Notes:- The synthesis protocol used herein was developed from existing protocols used by Vanzi et al. (2003) and van den Hout et al. (2011), and has been used in our recently published work (Beren et al., 2017).
- Polynucleotide phosphorylase from Synechocystis sp. is used to polymerize fluorescent polyU from UDP monomers, which is initiated by a short ‘seed’ oligo (500 pmol of a 5’ tagged cy5 polyU oligo 20-mer, but, in principle, this can be any length or sequence).
- Mix 500 pmol of seed oligos with 20 mM UDP, 1 μl of RNase Inhibitor and 300 ng of PNPase in Synthesis buffer in a total volume of 80 μl. Incubate at 37 °C to start the reaction.
- After a 2-h incubation at 37 °C, the UDP concentration is increased to a final concentration of 40 mM. Incubate the solution for another 2 h, for a total time of 4 h at 37 °C.
- Thoroughly wash the product three times with TE buffer on a 100-kDa Amicon filter using a tabletop centrifuge operated at 5,000 x g and 4 °C for 6 min.
- Determine the amount of polyU synthesized by measuring UV-Vis absorbance, and the absorbance ratio at 260 nm/280 nm is used as a measure of RNA purity (A260/A280 greater than 2.0).
- PolyU purification
- Fifty micrograms of fluorescent polyU per lane is electrophoresed on a 1% agarose/TAE RNA electrophoresis gel at room temperature and 90 V for 2 h alongside 2 outer lanes containing 7 μl of ssRNA ladder.
- Before loading on the gel, denature the ladder with formamide (70% of sample volume per lane) and heat to 70 °C for 10 min to eliminate all secondary structure.
Note: We found that formamide denaturation is not necessary for polyU RNA, as it already lacks secondary structure in the absence of formamide. Typically we run 7 μl of ssRNA ladder, and add 15 μl of formamide. - After disconnecting the power source, soak the gel in TAE buffer for 15 min, allowing the ladder to reform secondary structures and to be stained with ethidium bromide (or equivalent nucleic acid stain).
- Cut the fractions containing polyU of 500-1,500, 1,500-2,500, 2,500-3,500, 3,500-5,000, 5,000-7,000, and 7,000-9,000 nt from the gel and electroelute into dialysis bags with TAE buffer for 1 h at room temperature at 90 V. Briefly, excise a portion of the gel corresponding to the desired RNA length using a scalpel and place it inside a dialysis tube, which is then filled with the minimum volume of TAE needed to fully submerge the gel. Then place these tubes back into the electrophoresis tank and the desired RNA is eluted out of the gel and into the dialysis bag. It is important that during electroelution the gels are oriented to create the shortest possible path for the RNA to be eluted from the excised gel. (Video 1)Video 1. Gel electroelution protocol. This video illustrates the procedure of gel electroelution described above in Procedure C (PolyU Purification). The video illustrates Step C4, wherein the portion of the gel containing the desired RNA is excised and the RNA is then electroeluted out of this band into TAE buffer in dialysis tubing.
- After electroelution, wash the RNA three times with TE buffer using a 100-kDa Amicon filter, as described above. Determine the concentrations of polyU by UV-Vis spectrophotometry using the absorbance at 260 nm (using an extinction coefficient of 40 ng-cm/microliter). The purity of the electroeluted sample is subsequently verified by RNA gel electrophoresis.
- Determination of the size of polyU in solution
- Determine the hydrodynamic radii of three polyU RNA fractions (500-1,500, 1,000-2,500 and 2,500-3,500 nt) and three normal-composition RNAs (the 3,200-nt-long Brome Mosaic Virus (BMV) RNA1, and two truncations of it that are 500 and 2,000 nt long, respectively) by dynamic light scattering (DLS) (see Figure 3) (Hovanessian and Justesen, 2007).
- Perform these measurements on a Malvern particle sizer Nano-ZS with a sample concentration of 1 g/L in RNA Assembly Buffer (raw data are shown in Figure 3A). Radius against length in Figure 3B shows that the polyU radii exhibit N1/2 power-law scaling and that the normal-composition RNAs exhibit N1/3 power-law scaling, the scaling predicted for normal-composition RNAs by previous work using folding algorithms (Fang et al., 2011). In addition, the radii of polyU RNAs are approximately 25 percent larger than those of normal-compositions of similar length.
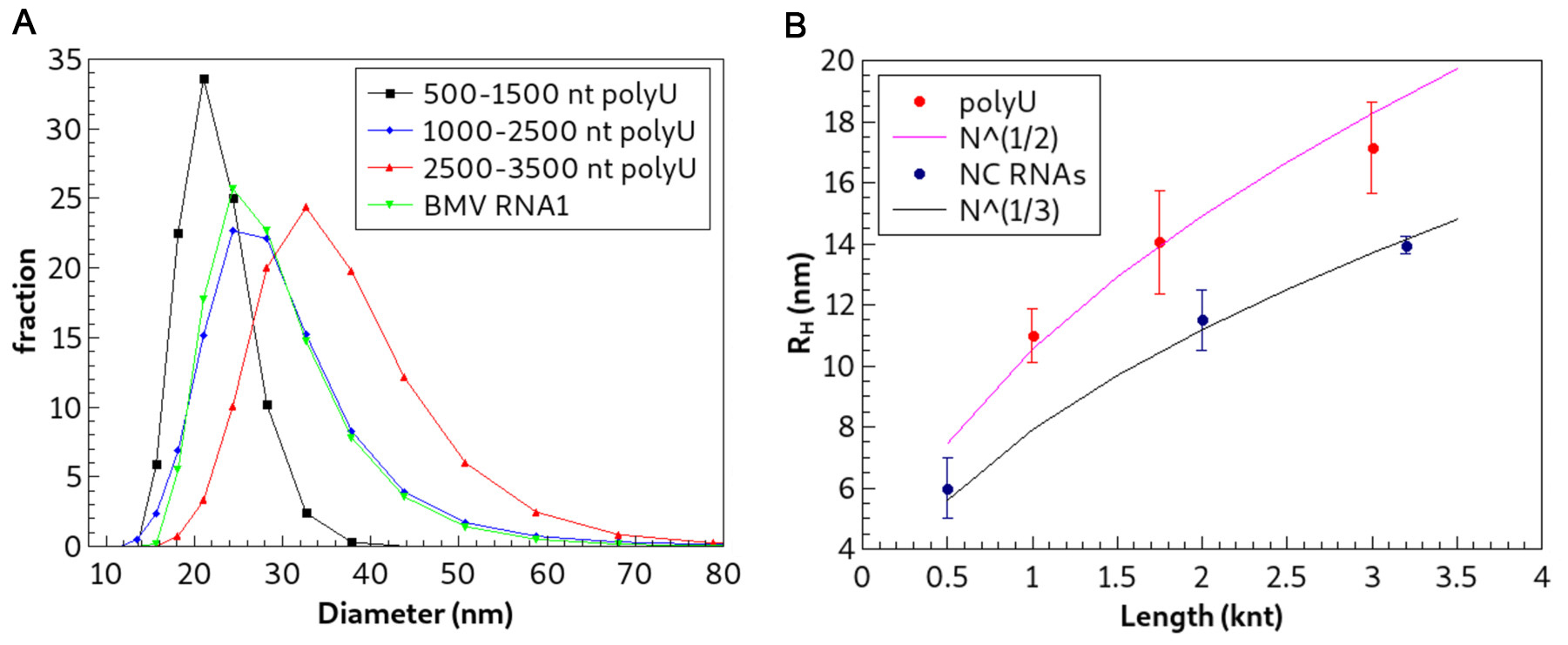
Figure 3. Dynamic light scattering measurements of the size of polyU in solution. A. Raw DLS data of polyU and BMV RNA 1 (3,200 nt long normal-composition RNA). The fraction of molecules of a given hydrodynamic diameter (nm) is plotted for each sample. The distributions are broad, but the average size persists between measurements. B. Plot showing how the hydrodynamic radii of polyU and normal-composition RNAs (500, 2,000 and 3,200 nt) vary with nt length. The magenta line that has been drawn through the data represents N1/2 power-law scaling (polyU), while the black line represents the N1/3 scaling expected for RNAs with secondary structure. Note that though the raw data exhibit a large standard deviation, the standard deviation of the mean (error bars) is relatively small, indicating that the average size is reproducible across measurements. (Adapted from Beren et al., 2017)
Notes
- The synthesis protocol is robust and has a similar yield and reproducibility to RNA transcription.
- We chose to produce polyU instead of polyA, polyG or polyC because while all of the RNA homopolymers lack the ability to self-base-pair, polyU also exhibits the weakest base-stacking, resulting in polyU having the least secondary structure of the four homopolymer RNAs.
- PNPase has the ability to synthesize polynucleotides without a template, and a synthesis of polyU using Alexa Fluor 488 fluorescent fUTP was carried out as well, resulting in viral-length RNAs that have a low-level of fluorescent UTP incorporation (Data Not Shown). However, this method of synthesis was not explored further in this work as it proved more difficult than the protocol described herein.
- Fractionation yields vary significantly from one experiment to another, with yields ranging from 20% to 70% of the RNA. This large variation results from excising narrow bands from agarose gels, with the end result being that there is significant variation in the lengths of polyU acquired between fractionations. Additionally, it is critical that everything be clean during the fractionation procedure to prevent degradation of the polyU RNA. Lastly, keeping the samples on ice as much as possible significantly improves the yields of the fractionation procedure by reducing RNA degradation.
Recipes
Notes:
- The following buffers are employed. All buffers are autoclaved and stored at 4 °C. All buffers use ddH2O from a Milli-Q UV Plus.
- *Adjust the buffers to the desired pHs using hydrochloric acid.
- Synthesis buffer
100 mM Tris base pH 9*
1 mM EDTA
5 mM MgCl2 - TE buffer
10 mM Tris base pH 7.5*
1 mM EDTA - TAE buffer
40 mM Tris base pH 8*
20 mM acetic acid
1 mM EDTA - RNA assembly buffer
50 mM Tris base pH 7.2*
50 mM NaCl
10 mM KCl
5 mM MgCl2
Acknowledgments
Author contributions: All authors were involved in designing the experiments. C.B., L.D. and K.N.L. performed the experiments and analyzed the data. C.B., W.G. and C.K. wrote this article together.
This protocol was adapted from Beren et al. (2017).
This research was supported by grant CHE1051507 from the NSF. KNL was supported by the Amgen Scholars Program over the course of this work.
Competing interests
The authors declare no conflicts of interest.
References
- Beren, C., Dreesens, L. L., Liu, K. N., Knobler, C. M. and Gelbart, W. M. (2017). The effect of RNA secondary structure on the self-assembly of viral capsids. Biophys J 113(2): 339-347.
- Fang, L. T., Gelbart, W. M. and Ben-Shaul, A. (2011). The size of RNA as an ideal branched polymer. J Chem Phys 135(15): 155105.
- Garmann, R. F., Gopal, A., Athavale, S. S., Knobler, C. M., Gelbart, W. M. and Harvey, S. C. (2015). Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA 21(5): 877-886.
- Gopal, A., Zhou, Z. H., Knobler, C. M. and Gelbart, W. M. (2012). Visualizing large RNA molecules in solution. RNA 18(2): 284-299.
- Grunberg-Manago, M., Ortiz, P. J. and Ochoa, S. (1956). Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of Azotobacter vinelandii. Biochim Biophys Acta 20(1): 269-285.
- Hovanessian, A. G. and Justesen, J. (2007). The human 2'-5'oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2'-5' instead of 3'-5' phosphodiester bond formation. Biochimie 89(6-7): 779-788.
- Inners, L. D. and Felsenfeld, G. (1970). Conformation of polyribouridylic acid in solution. J Mol Biol 50(2): 373-389.
- Martin, R. G. and Ames, B. N. (1962). The effect of polyamines and of poly U size on phenylalanine incorporation. Proc Natl Acad Sci U S A 48(12): 2171-2178.
- Richards, E. G., Flessel, C. P. and Fresco, J. R. (1963). Polynucleotides. VI. Molecular properties and conformation of polyribouridylic acid. Biopolymers 1(5):431-446.
- Vanzi, F., Vladimirov, S., Knudsen, C. R., Goldman, Y. E. and Cooperman, B. S. (2003). Protein synthesis by single ribosomes. RNA 9(10): 1174-1179.
- van den Hout, M., Skinner, G. M., Klijnhout, S., Krudde, V. and Dekker, N. H. (2011). The passage of homopolymeric RNA through small solid-state nanopores. Small 7(15): 2217-2224.
- Yoffe, A. M., Prinsen, P., Gopal, A., Knobler, C. M., Gelbart, W. M. and Ben-Shaul, A. (2008). Predicting the sizes of large RNA molecules. Proc Natl Acad Sci U S A 105(42): 16153-16158.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Beren, C., Liu, K. N., Dreesens, L. L., Knobler, C. M. and Gelbart, W. M. (2018). Enzymatic Synthesis and Fractionation of Fluorescent PolyU RNAs. Bio-protocol 8(17): e2988. DOI: 10.21769/BioProtoc.2988.
Category
Molecular Biology > RNA > RNA labeling
Biochemistry > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link