- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Two Different Methods of Quantification of Oxidized Nicotinamide Adenine Dinucleotide (NAD+) and Reduced Nicotinamide Adenine Dinucleotide (NADH) Intracellular Levels: Enzymatic Coupled Cycling Assay and Ultra-performance Liquid Chromatography (UPLC)-Mass Spectrometry
(*contributed equally to this work) Published: Vol 8, Iss 14, Jul 20, 2018 DOI: 10.21769/BioProtoc.2937 Views: 8935
Reviewed by: Andrea PuharNeelanjan BoseAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of Chloroplastic NAD Kinase Activity and Whole Tissue NAD Kinase Assay
Yuuma Ishikawa [...] Shin-nosuke Hashida
Jan 5, 2020 4879 Views

Quantitative Analysis of Redox Pool (NAD+, NADH Content) in Plant Samples Under Aluminum Stress
Jay Prakash Awasthi [...] Sanjib Kumar Panda
Jun 20, 2022 3339 Views
Abstract
Current studies on the age-related development of metabolic dysfunction and frailty are each day in more evidence. It is known, as aging progresses, nicotinamide adenine dinucleotide (NAD+) levels decrease in an expected physiological process. Recent studies have shown that a reduction in NAD+ is a key factor for the development of age-associated metabolic decline. Increased NAD+ levels in vivo results in activation of pro-longevity and health span-related factors. Also, it improves several physiological and metabolic parameters of aging, including muscle function, exercise capacity, glucose tolerance, and cardiac function in mouse models of natural and accelerated aging.
Given the importance of monitoring cellular NAD+ and NADH levels, it is crucial to have a trustful method to do so. This protocol has the purpose of describing the NAD+ and NADH extraction from tissues and cells in an efficient and widely applicable assay as well as its graphic and quantitative analysis.
Background
Oxidized Nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide (NAD+ and NADH) are important biological cofactors that donate and accept electrons in several anabolic and catabolic functions. They participate in reactions such as glycolysis, the tricarboxylic acid cycle, and oxidative phosphorylation. In addition, it serves as a substrate for several enzymes involved in DNA damage repair, such as the sirtuins and poly (ADP-ribose) polymerases (PARPs) (Imai and Guarente, 2014; Verdin, 2015; Yoshino et al., 2018).
NAD+ levels decrease during aging and are involved in age-related metabolic decline. It has been shown that the cellular NAD pool is determined by a balance between the activity of NAD-synthesizing and NAD-consuming enzymes (Aksoy et al., 2006; Barbosa et al., 2007; Yang et al., 2007; Nahimana et al., 2009; Bai et al., 2011; Yoshino et al., 2011). In previous publications, our laboratory has demonstrated that expression and activity of the NADase CD38 increases with age and that CD38 is required for the age-related NAD decline and mitochondrial dysfunction via a pathway mediated at least in part by regulation of SIRT3 activity (Camacho-Pereira et al., 2016). We also identified CD38 as the main enzyme involved in the degradation of the NAD precursor nicotinamide mononucleotide (NMN) in vivo. That indicates that CD38 has a key role in the modulation of NAD-replacement therapy for aging and metabolic diseases (Camacho-Pereira et al., 2016). CD38 was originally identified as a cell-surface enzyme that plays a key role in several physiological processes such as immune response, inflammation, cancer, and metabolic disease (Frasca et al., 2006; Barbosa et al., 2007; Guedes et al., 2008; Malavasi et al., 2008).
Several different assays and methods have been described due to the great importance of monitoring NAD+ and NADH cellular levels under various physiological conditions. Specifically two of them, ultra-performance liquid chromatography (UPLC)-mass spectroscopy assay and cycling assay have finality and both are equally sensitive and specific (Camacho-Pereira et al., 2016).
Our laboratory also further optimized and validated the cycling assay. We determined NAD+ and NADH specific and does not detect any of the other nucleotides or NAD derivatives tested, including nicotinamide adenine dinucleotide phosphate (NADP), nicotinic acid adenine dinucleotide (NAAD), nicotinic acid adenine dinucleotide phosphate (NAADP), cyclic-adenine diphosphate ribose (cADPR), adenine triphosphate (ATP), ADP, and others. The results obtained with both methods confirm that there is indeed a decrease in levels of both NAD+ and NADH in murine tissues during chronological aging (Camacho-Pereira et al., 2016). Furthermore, both techniques correlated well for both nucleotides (correlation coefficient of r = 0.95 for NAD+ and 0.97 for NADH) (Camacho-Pereira et al., 2016).
In regards to the cycling assay (pathway in which the main reaction happens), it takes NAD+ and NADH present in the samples and they are coupled to both enzymes alcohol dehydrogenase (ADH) and diaphorase in a cycling assay. Every time NAD+ or NADH cycles, it produces a molecule of resorufin, which is highly fluorescent. This fluorescence is directly captured by the multi-well fluorescence plate reader therefore indirectly the NAD+ levels can be measured. Most studies have relied on separated extractions for NAD+ and reduced nicotinamide adenine dinucleotide (NADH) determination: a basic extraction for the reduced species and a separate acidic extraction for the oxidized species (Ashrafi et al., 2000; Smith et al., 2000; Lin et al., 2001; Anderson et al., 2002; Lin et al., 2004). The extraction conditions are specific for the stabilization of either oxidized compounds, which are more stable in acid or reduced compounds, which are more stable in base.
Conversely, the ultra-performance liquid chromatography (UPLC)-mass spectrometry is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography with the mass analysis capabilities of mass spectrometry (MS). MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides identity of the individual components with high molecular specificity and detection sensitivity. This tandem technique can be used to analyze very accurate components such as NAD+ and NADH (Dass, 2007).
Part I: Cycling assay
Materials and Reagents
- Plastic tips 10 (Thermo Fisher Scientific, Molecular Bioproducts, catalog number: 3511-05 )
- Plastic tips 1,000 (Thermo Fisher Scientific, Molecular Bioproducts, catalog number: 3101 )
- Plastic tips 200 (Thermo Fisher Scientific, Molecular Bioproducts, catalog number: 3551 )
- 1.5 ml Microcentrifuge tubes (USA Scientific, catalog number: 1415-2508 )
- 2.0 ml MCT Graduated tubes (Fisher Scientific, Fisherbrand, catalog number: 05-408-146 )
- 96-well Microfluor 1White flat-bottom plate (Thermo Fisher Scientific, catalog number: 7705 )
- Combitips Advanced 10 ml (Eppendorf, catalog number: 0030089464 )
- Disposable cell lifter (Fisher Scientific, Fisherbrand, catalog number: 08-100-240 )
- Cells of interest: any cell can be used to measure NAD+/NADH levels
- Tissues of interest: any tissue can be used to measure NAD+/NADH levels
- Trichloroacetic Acid (TCA) (Sigma-Aldrich, catalog number: T4885-500G )
- Sodium hydroxide (NaOH) 10 N (Fisher Scientific, Fisher Chemical, catalog number: SS267 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- 1,2,2-Trichlorotrifluoroethane (TCTFE) (Fisher Scientific, Fisher Chemical, catalog number: T1781 )
- Trioctylamine (Sigma-Aldrich, catalog number: T81000-500G )
- Tris base, Trizma base (Sigma-Aldrich, catalog number: T6066 )
- Diaphorase (Sigma-Aldrich, catalog number: D5540-500UN )
- Alcohol dehydrogenase (ADH) (Sigma-Aldrich, catalog number: A3263-75KU )
- Activated charcoal (Sigma-Aldrich, catalog number: C7606-125G )
- Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: S0751-500G )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S0876-500G )
- Absolute Ethanol (Fisher Scientific, Fisher BioReagentsTM, catalog number: BP2818-500 )
- Bovine Serum Albumin (Sigma-Aldrich, catalog number: A7906 )
- β-NAD sodium salt (Sigma-Aldrich, catalog number: N0632-1G )
- Riboflavin 5’-monophosphate sodium salt hydrate (FMN) (Sigma-Aldrich, catalog number: F8399 )
- Resazurin sodium salt (Sigma-Aldrich, catalog number: 199303-5G )
- Phosphate-buffered saline (PBS – pH 7.4) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Organic solvent (see Recipes)
- Sodium Phosphate Buffer (pH 8.0) (see Recipes)
- 20 mM Sodium Phosphate Buffer (pH 7.0) (see Recipes)
- 4% charcoal suspension (in 20 mM sodium phosphate, pH 7.0) (see Recipes)
- Bradford Dye (see Recipes)
- Tris Buffer (1 M, pH 8.0) (see Recipes)
Equipment
Note: The brands and models indicated are the ones used by our group, similar equipment can be used as well.
- 1 L volumetric flask
- Pipettes (10, 200, 1,000 µl)
- Repeat Pipette (Eppendorf, model: Repeater® M4 )
- Scissors
- Homogenizer, Tissue Tearor (Bio Spec Products, catalog number: 780CL-04 )
- Microcentrifuge (Eppendorf, model: 5424 )
- Scale (Mettler-Toledo International, model: AG104 )
- Sonic Dismembrator (Fisher Scientific, model: Model 100 )
- Vortex (Scientific Industries, model: Vortex-Genie 2 , catalog number: G560)
- -80 °C freezer
- Plate reader (Molecular Devices, model: SpectraMax® GeminiTM XPS )
- Spectrophotometer (BioTek Instruments, model: Epoch 2 )
Software
- Gen5 Microplate Reader and Imager Software (BioTek Instruments)
- Microsoft Excel (Microsoft Corporation)
- SoftMax Pro 6 (Molecular Devices, LLC)
- GraphPad Prism 7 (GraphPad Software, Inc)
Procedure
- Sample preparation
If using cells, use at least 3 x 106 cells. Aspirate media from plate, wash with PBS, collect the cells scratching the plate using disposable cell lifter, and pellet by centrifugation (30 sec, at 2,000 x g). Aspirate supernatant, and proceed to Step A1. If measuring NAD+ from tissue samples cut approximately 20 mg of tissue and put the piece in a 2 ml tube. Then proceed to Step A1. For all the following steps, samples should be kept on ice.
Considering either NAD+ or NADH quantification, the only step that distinguishes those two is the extraction part. From Step A2 on, the process is the same for both NAD+ and NADH.- Extraction:
- For NAD+: start extraction of NAD+ by adding 10% TCA (diluted in water).
- For NADH: NADH is extracted with 500 mM NaOH and 5 mM EDTA.
- For NAD+: start extraction of NAD+ by adding 10% TCA (diluted in water).
- Homogenize tissue samples with scissors and mechanical homogenizer until there are no visible chunks. Keep samples always on ice during this process.
Note: Skip this step if using cells. - Sonicate samples 3 times for 5 sec each. Configure equipment on medium power.
Note: For NADH: at this point, heat for 30 min at 60 °C. - Pellet protein by centrifuging at 11,500 x g for 2 min, 4 °C. Transfer supernatant to a new 1.5 ml tube and use it for Step A6.
- Save pellet for later protein determination. Re-suspend pellet in NaOH:
- Cells: 0.2 M NaOH, volume based on pellet size; around 100 µl (always better to use a lower volume and dilute the protein sample after, than to use a larger volume and dilute it too much). Samples should be kept on ice or a rocker platform at 4 °C until it is completely soluble.
- Tissue: 1 M NaOH, at least 300 µl. Leave samples on a rocker at 4 °C overnight and then measure protein concentration.
- We measure the protein concentration by the Bradford protein assay.
- Cells: 0.2 M NaOH, volume based on pellet size; around 100 µl (always better to use a lower volume and dilute the protein sample after, than to use a larger volume and dilute it too much). Samples should be kept on ice or a rocker platform at 4 °C until it is completely soluble.
- Remove TCA using organic solvent
- Organic solvent: prepare a fresh solution of TCTFE:Trioctylamine (3:1 ratio). Use only plastic tips and tubes (TCTFE = 1,1,2-trichloro-1,2,2-trifluoroethane).
- Add organic solvent to remove TCA (supernatant of Step A4) at a ratio of 2:1.
- Vortex vigorously for 15 sec allowing mixture to separate at room temperature for 3-5 min.
- Carefully remove the top aqueous layer containing the NAD+ (100 µl) with a plastic pipette (Figure 1), put it in a new 1.5 ml tube, and add 10 µl of 1 M Tris to adjust the pH to 8.0.
Note: If by mistake the bottom organic layer is perturbed or aspirated, put it back, and repeat Steps A6c and A6d. - Put samples on ice and proceed to enzyme preparing steps. At this point, samples can be frozen in a -80 °C freezer and procedure can be resumed at another time, although it is advised to proceed with the procedure and reading, if possible, to minimize risk of degradation in freeze-thaw cycles.

Figure 1. Removing top aqueous layer
- Organic solvent: prepare a fresh solution of TCTFE:Trioctylamine (3:1 ratio). Use only plastic tips and tubes (TCTFE = 1,1,2-trichloro-1,2,2-trifluoroethane).
- At this step, dilute NAD+ samples in sodium phosphate buffer considering the range of the standard curve. For all the samples NAD+/NADH measurement has to be within the curve. The dilution varies depending on the type and size of tissues or cells. It is basically empirical. Usually for 20 mg tissues, the dilutions range from 1:500 to 1:1,000. For cells, the dilution usually ranges from 1:50 to 1:200. Remember this is not rigid; sometimes a lower or higher dilution is needed. Dilute NAD+ samples, to a final volume of at least 250 µl.
- Extraction:
- Preparing NAD+ standard curve
- In order to obtain a final NAD+ 1 µM concentration, it is necessary to perform a serial dilution as indicated below:
- In a 1.5 ml microcentrifuge tube, weigh β-NAD sodium salt (around 1 mg).
- Using the β-NAD sodium salt molecular weight find the specific volume to get a solution of 10 mM.
- To get the intermediate concentration of 10 µM, dilute the 10 mM 1:1,000 in sodium phosphate buffer.
- Finally, to get the solution concentration of 1 µM, dilute the 10 µM 1:10 in sodium phosphate buffer.
- In a 1.5 ml microcentrifuge tube, weigh β-NAD sodium salt (around 1 mg).
- To obtain the standard curve, dilute the 1 µM stock of NAD+ in sodium phosphate buffer following Table 1 below.
Table 1. NAD+ Standard Curve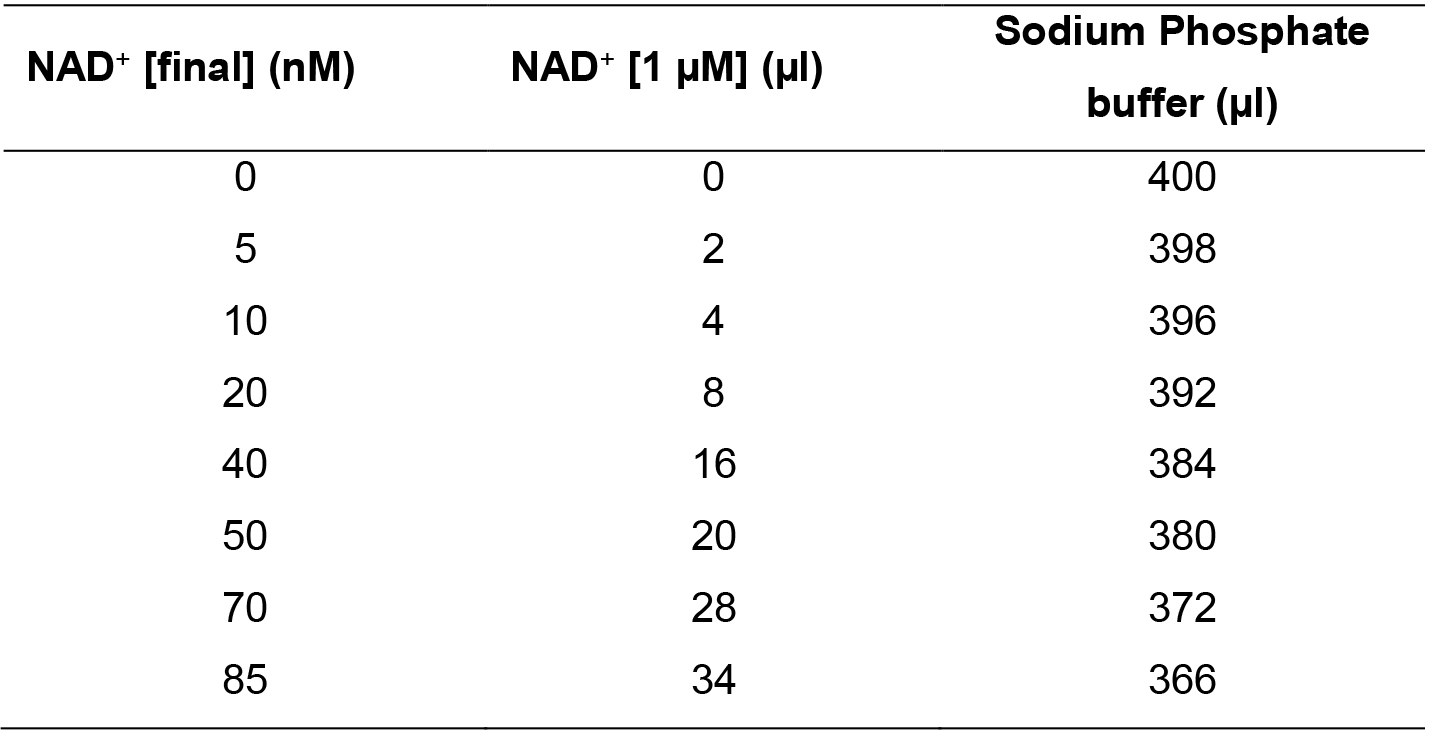
- In order to obtain a final NAD+ 1 µM concentration, it is necessary to perform a serial dilution as indicated below:
- Preparing the enzymes
- Based on Table 2:
Stock for ADH has to be 3400 U/ml.
Stock for Diaphorase has to be 100 U/ml. - Weigh the enzymes in a 1.5 ml tube for preparation of 1 ml solution. Check the amount of U/mg in the bottle being used as it might change depending on the lot number.
Notes:- Usually, we prepare 1 ml of each enzyme solution and divide in smaller aliquots in a size enough to prepare at least 1 plate (96 wells), in order to avoid thawing the whole amount each time.
- We suggest preparing approximately 3-4% more volume (100 wells instead of 96 wells for entire plate), considering that some volume can be lost when moving the solution to another tube (e.g., attached to the tube walls, to the pipet tips).
- Usually, we prepare 1 ml of each enzyme solution and divide in smaller aliquots in a size enough to prepare at least 1 plate (96 wells), in order to avoid thawing the whole amount each time.
- Remove bound NAD+ from Diaphorase and ADH by diluting the enzymes in 30% (300 µl) of sodium phosphate buffer and 70% (700 µl) of 4% charcoal suspension for a total of 1 ml solution. Gently mix the solution, and incubate it for 40 min at 37 °C. Mix gently every 10 min.
- Remove charcoal by centrifuging for 5 min, 4 °C, and 13,500 x g. Transfer supernatant to a new tube and centrifuge for 2 min, 4 °C, and 13,500 x g.
- Based on Table 2:
- Preparing the reaction mix
- Thaw FMN on ice, and bring it to room temperature.
- Whenever possible, prepare a Resazurin stock of 20 mM in water and use it on the same day (protect it from light until use).
Note: According to manufacturer’s instructions, small aliquots of the Resazurin stock can be stored at -20 °C for up to 1 month (stock solutions should be protected from exposure to light). When needed to be used, thaw it on ice. - Prepare the reaction mix following Table 2 below, respecting the amount and the order in which each component has to be added.
Table 2. Reaction mix preparation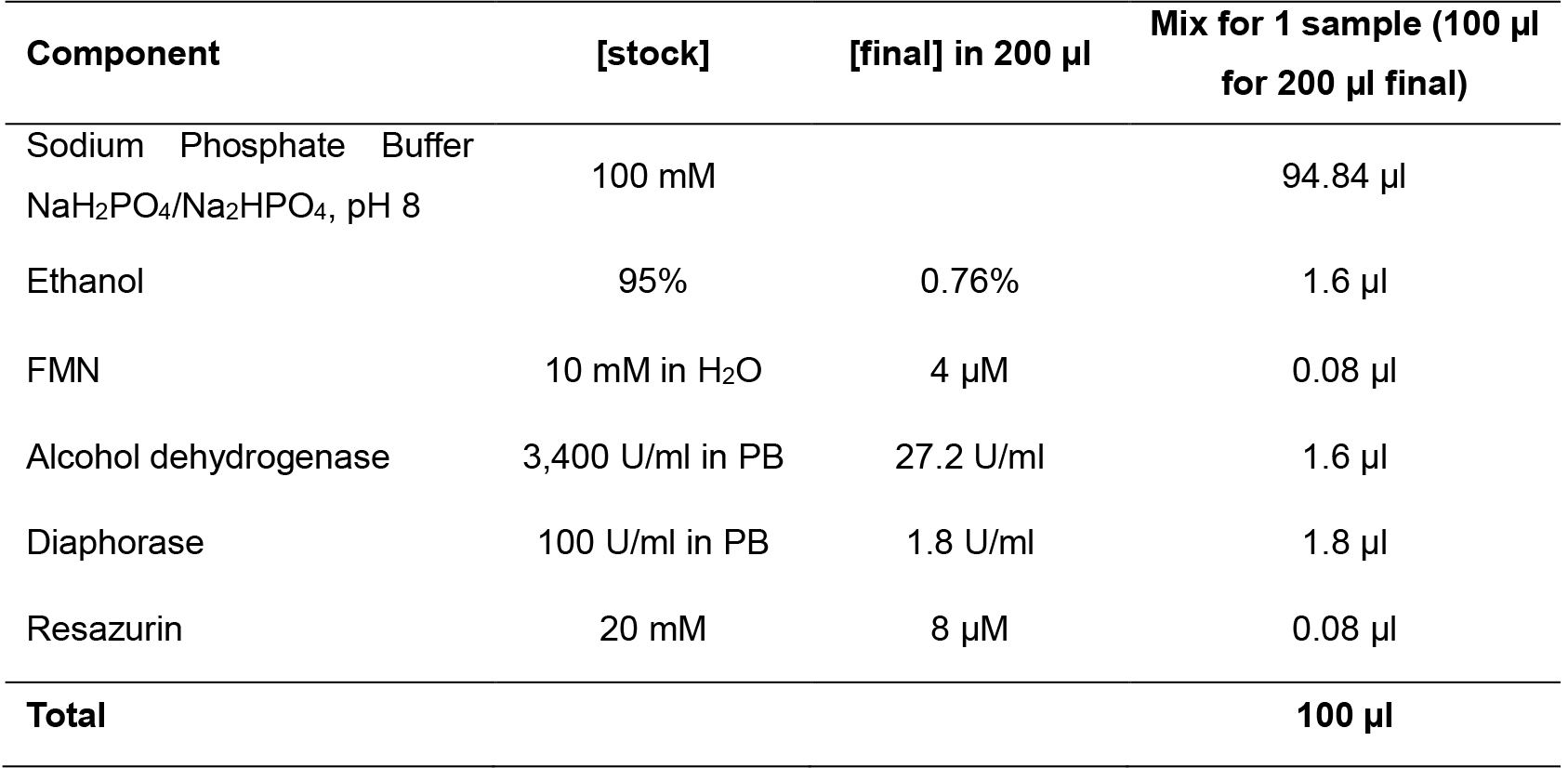
- Thaw FMN on ice, and bring it to room temperature.
- Setting up the plate
- Add 100 µl of sample/standards into wells of a 96-well opaque plate.
- Add 100 µl of reaction mix to each well with a repeat pipet. Put the 96-well plate into a fluorescence plate reader. Monitor for 60 min with an excitation wavelength of 544 nm and an emission wavelength of 590 nm (Video 1).Video 1. Setting up the plate reader
- On template editor, configure dilution and unit for NAD+ Standards, and dilution of samples (Figure 2).
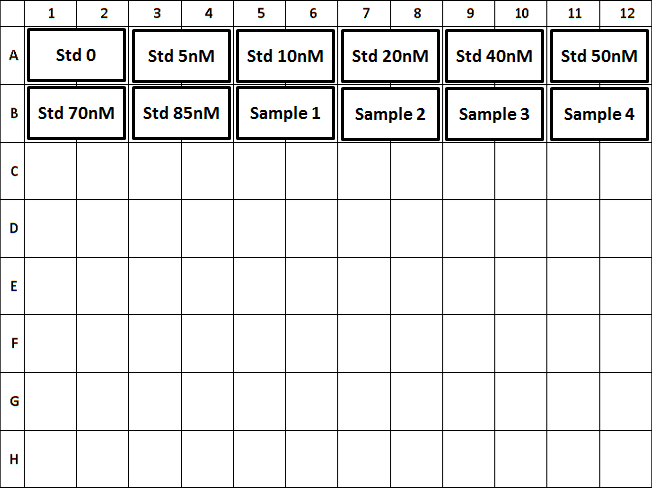
Figure 2. Plate layout
- Add 100 µl of sample/standards into wells of a 96-well opaque plate.
Data analysis
- Data needed: NAD+ values obtained in fluorescence plate reader (Figure 3A), protein concentration, volume of TCA 10% and NaOH used.
- Calculate NAD+ concentration in nmol by dividing the NAD+ value obtained (in µM) by the volume of TCA (in ml).
- Calculate the mass of protein (mg) present in the sample by dividing protein concentration (mg/ml) by volume of NaOH (ml) in which the pellet was re-suspended.
- Divide the NAD+ concentration obtained in Step 2 by the mass of protein obtained in Step 3. This is the final result (Figure 3B).
- Plot the results (nM/mg protein) in a graph.
- Perform statistical analysis as needed.
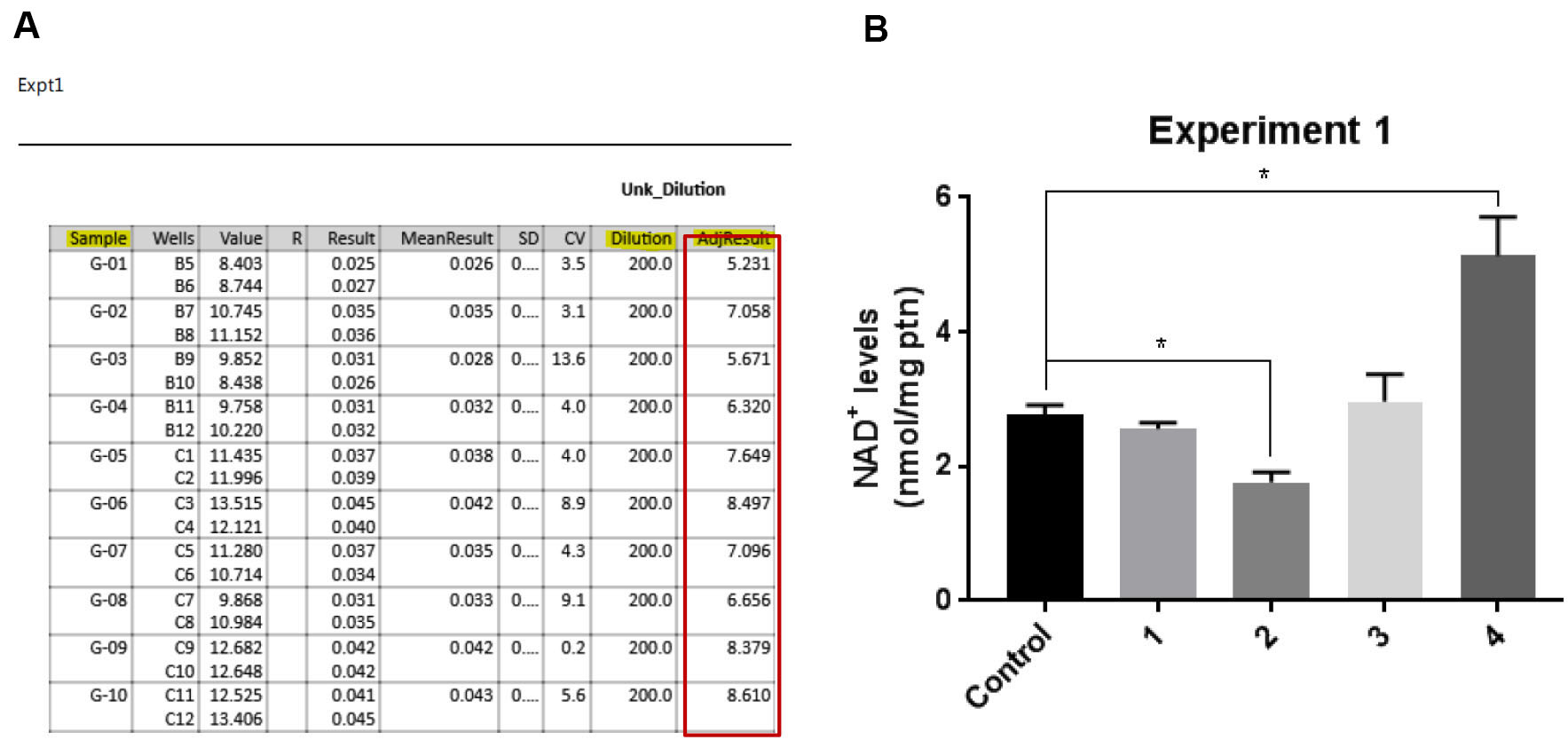
Figure 3. Data analysis. A. Raw data from plate reader; the encircled detail shows the adjusted result based on dilution. B. Example of final graph.
Recipes
- Organic solvent
Prepare a fresh solution of TCTFE:Trioctylamine (3:1 ratio)
Use only plastic tips and tubes (TCTFE = 1,1,2-trichloro-1,2,2-trifluoroethane) - 100 mM Sodium Phosphate Buffer (pH 8.0)
In a 1 L volumetric flask:
Add 93.2 ml of 1 M Na2HPO4 followed by 6.6 ml of 1 M NaH2PO4
Bring the volume up to 1 L
The buffer can be stored up to 1 month at 4 °C - 20 mM Sodium Phosphate Buffer (pH 7.0)
Use the 100 mM Sodium Phosphate Buffer (pH 8.0) to make a 1:5 dilution
For a final volume of 500 ml, add 100 ml of 100 mM Sodium Phosphate Buffer (pH 8.0) to a beaker
Adjust to pH 7.0 using 1 N HCl solution
Bring the volume up to 500 ml using a graduated cylinder
Store at room temperature - 4% charcoal suspension (in 20 mM sodium phosphate, pH 7)
Weigh 4 g of charcoal
Transfer it to a graduated cylinder and add 100 ml of 20 mM Sodium Phosphate Buffer (pH 7.0)
Store at room temperature - Bradford Dye
Dilute Bio-Rad Protein Assay Dye Reagent Concentrate 1:5 in water
The diluted reagent may be used for approximately 2 weeks when kept at room temperature - Tris Buffer (1 M, pH 8.0)
Put 8 ml of distilled water in a suitable container
Add 1.21 g of Tris base to the solution
Adjust the solution pH to 8.0 with HCl
Add distilled water until the volume is 10 ml
Part II: Ultra-performance liquid chromatography (UPLC)-mass spectrometry
Materials and Reagents
- Agilent Poroshell 120 EC-C18 pre-column (Agilent Technologies, catalog number: 693775-902 )
- Agilent Poroshell 120 EC-C18 analytical column (Agilent Technologies, catalog number: 697975-902T )
- Formic acid (Fisher Scientific, catalog number: A117-50 )
- Acetonitrile (Sigma-Aldrich, catalog number: 271004 )
- NAD (Sigma-Aldrich, catalog number: N3014 )
- NADH (Sigma-Aldrich, catalog number: N6005 )
- n-cyclohexyl benzamide (Internal Standard - IS) (Sigma-Aldrich, catalog number: R531332 )
- Auto-sampler vials (Chrom Tech, catalog number: CTI-9405 )
- Slick microfuge tubes (Fisher Scientific, FisherbrandTM, catalog number: 02-681-320 )
- Primary stock solutions (see Recipes)
Equipment
- LC-MS/MS system
Note: The LC-MS system consists of a Waters ACQUITY H class ultra-performance liquid chromatography (UPLC) system, containing a quaternary solvent manager and sample manager-FTN coupled to a Xevo TQ-S mass spectrometer (Waters, Milford, MA) equipped with electrospray ionization (ESI) source.
Software
- Waters MassLynx v4.1 software
Procedure
- The liquid chromatographic separation of NADH and NAD+ is accomplished using an Agilent Poroshell 120 EC-C18 pre-column (2.1 x 5 mm, 2.7 μm, Chrom Tech, Apple Valley, MN) attached to an Agilent Poroshell 120 EC-C18 analytical column (2.1 x 100 mm, 2.7 μm, Chrom Tech, Apple Valley, MN) at 40 °C.
- Elute the column with a gradient mobile phase composed of water with 0.1% formic acid (A) and acetonitrile (ACN) with 0.1% formic acid (B) with a constant flow rate of 0.5 ml/min and a total run time of 10 min. The elution is initiated at 100% A and held for 2 min, then B is linearly increased from 0% to 60% B for 4 min, 60% B is held for 1 min, and returning to initial conditions in 2 min, finishing with 2.8 min at 100% A.
- Auto-sampler temperature is 4 °C and sample injection volume is 10 μl. NAD+ and NADH should be eluted for 1.6 and 3.5 min, respectively. None of the other NAD+ related metabolites (including NADP, NADPH, ATP, NAADP, NAAD, ADP, cADPR, ADPR, nicotinamide and nicotinic acid) are co-eluted with either NAD+ or NADH.
- Tissue samples are prepared as the same manner as described in “sample preparation” for the cycling assay. Samples should be thawed on ice and they are diluted in water containing 500 ng/ml of internal standard.
- Samples for NAD+ measurement diluted 1:1,000 and 1:10,000, whereas the ones for NADH samples are diluted 1:1,000 in slick microfuge tubes. After dilution, the samples should be vortexed and transferred to 2 ml auto-sampler vials for immediate analysis.
Data analysis
Data were acquired and analyzed by Waters MassLynx v4.1 software. Detection of NAD+ and NADH is accomplished using the mass spectrometer in positive ESI mode using capillary voltage 3.5 kV, source temp 150 °C, desolvation temp 500 °C, cone gas flow 150 L/h, desolvation gas flow 500 L/h, using multiple reaction monitoring (MRM) scan mode with a dwell time of 0.075 sec (Figure 4). The cone voltages and collision energies were determined by MassLynx- Intellistart, v4.1, software. Values are as follows:
NAD+–m/z 664.27 > 136.09–cone voltage 54 V–Collision 42 eV
NADH–m/z 666.28 > 514.17–cone voltage 56 V–Collision 26 eV
n-Cyclohexyl benzamide–m/z 204.1 > 122.00–Cone voltage 80 V–Collision 20 eV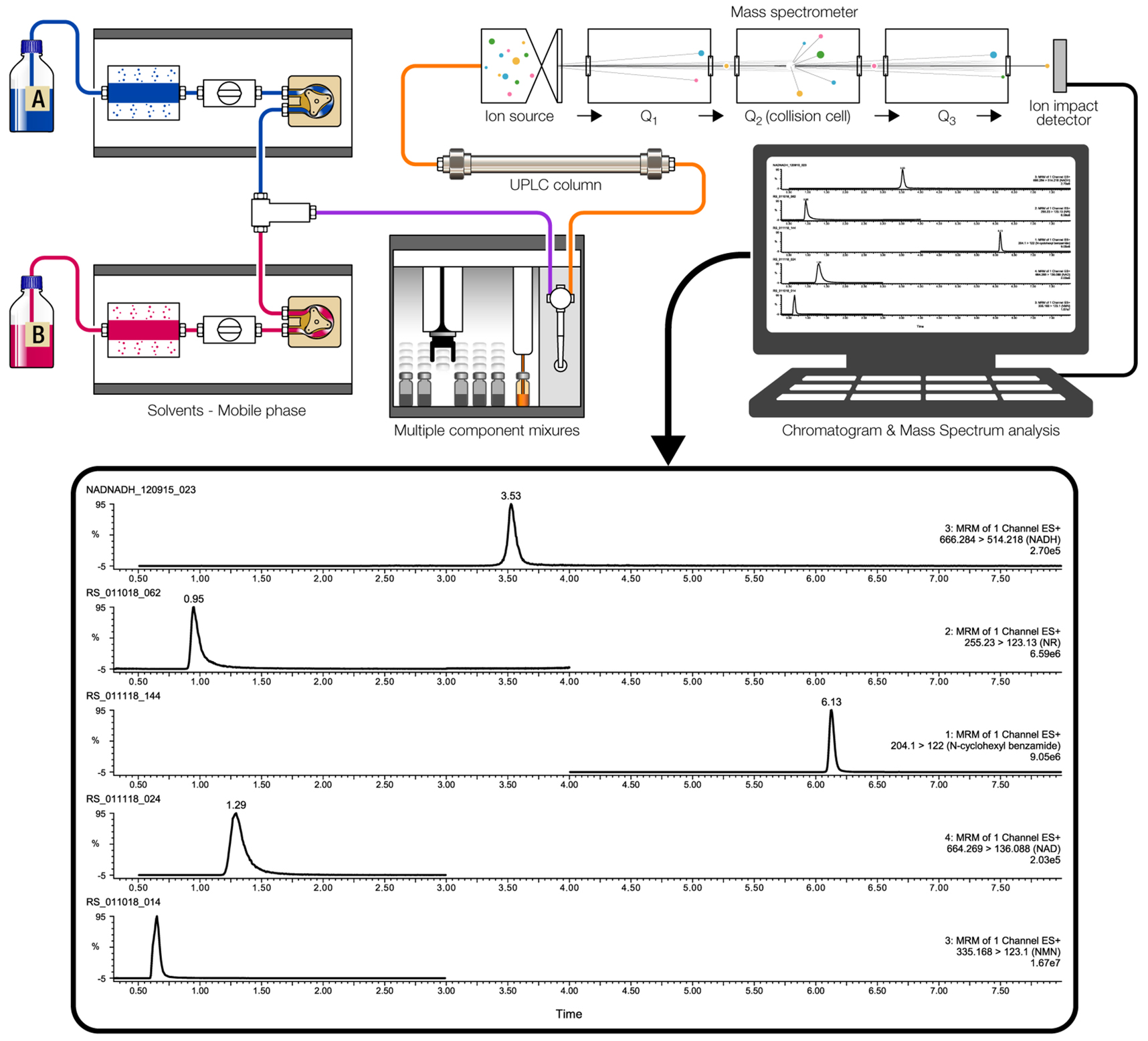
Figure 4. LC-MS tracing analysis
Recipes
- Primary stock solutions
The primary stock solutions were stored at -80 °C and were prepared in silanized glass vials as follows:
NAD+ (10 mg/ml in water)
NADH (1 mg/ml in 0.01 N NaOH, pH 11.9)
n-cyclohexyl benzamide (IS) (100 µg/ml in EtOH)
For working standards:
Working standards were prepared by dilution of the stock solution into the same solutions mentioned above. Daily standard samples were prepared by diluting above stocks 1:20 in water containing 500 ng/ml internal standard
Acknowledgments
This work was supported in part by grants from the Ted Nash Long Life Foundation, the Glenn Foundation for Medical Research via the Paul F. Glenn Laboratories for the Biology of Aging at the Mayo Clinic, a grant from Calico Laboratories, the Mayo Foundation, National Institutes of Health (NIH) grants from the National Institute of Aging (NIA, grant No. AG-26094), the Pancreatic Cancer SPORE project from NIH/NCI to E.N.C (grant No. CA102701-08), the Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567). Maria Auxiliadora-Martins was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Brazil (#2016/20089-4).
Declaration of interests: Dr. Chini holds a patent on the use of CD38 inhibitors for metabolic diseases.
References
- Aksoy, P., White, T. A., Thompson, M. and Chini, E. N. (2006). Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun 345(4): 1386-1392.
- Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., Cohen, H., Lin, S. S., Manchester, J. K., Gordon, J. I. and Sinclair, D. A. (2002). Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem 277(21): 18881-18890.
- Ashrafi, K., Lin, S. S., Manchester, J. K. and Gordon, J. I. (2000). Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev 14(15): 1872-1885.
- Bai, P., Canto, C., Oudart, H., Brunyanszki, A., Cen, Y., Thomas, C., Yamamoto, H., Huber, A., Kiss, B., Houtkooper, R. H., Schoonjans, K., Schreiber, V., Sauve, A. A., Menissier-de Murcia, J. and Auwerx, J. (2011). PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13(4): 461-468.
- Barbosa, M. T., Soares, S. M., Novak, C. M., Sinclair, D., Levine, J. A., Aksoy, P. and Chini, E. N. (2007). The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J 21(13): 3629-3639.
- Camacho-Pereira, J., Tarrago, M. G., Chini, C. C. S., Nin, V., Escande, C., Warner, G. M., Puranik, A. S., Schoon, R. A., Reid, J. M., Galina, A. and Chini, E. N. (2016). CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab 23(6): 1127-1139.
- Dass, C. (2007). Chap. 5. Hyphenated separation techniques. Fundamentals of contemporary mass spectrometry, 1. John Wiley & Sons, Inc., 151-194.
- Frasca, L., Fedele, G., Deaglio, S., Capuano, C., Palazzo, R., Vaisitti, T., Malavasi, F. and Ausiello, C. M. (2006). CD38 orchestrates migration, survival, and Th1 immune response of human mature dendritic cells. Blood 107(6): 2392-2399.
- Guedes, A. G., Jude, J. A., Paulin, J., Kita, H., Lund, F. E. and Kannan, M. S. (2008). Role of CD38 in TNF-α-induced airway hyperresponsiveness. Am J Physiol Lung Cell Mol Physiol 294(2): L290-299.
- Imai, S. and Guarente, L. (2014). NAD+ and sirtuins in aging and disease. Trends Cell Biol 24(8): 464-471.
- Lin, S. J., Ford, E., Haigis, M., Liszt, G. and Guarente, L. (2004). Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18(1): 12-16.
- Lin, S. S., Manchester, J. K. and Gordon, J. I. (2001). Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem 276(38): 36000-36007.
- Malavasi, F., Deaglio, S., Funaro, A., Ferrero, E., Horenstein, A. L., Ortolan, E., Vaisitti, T. and Aydin, S. (2008). Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev 88(3): 841-886.
- Nahimana, A., Attinger, A., Aubry, D., Greaney, P., Ireson, C., Thougaard, A. V., Tjornelund, J., Dawson, K. M., Dupuis, M. and Duchosal, M. A. (2009). The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood 113(14): 3276-3286.
- Smith, J. S., Brachmann, C. B., Celic, I., Kenna, M. A., Muhammad, S., Starai, V. J., Avalos, J. L., Escalante-Semerena, J. C., Grubmeyer, C., Wolberger, C. and Boeke, J. D. (2000). A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A 97(12): 6658-6663.
- Verdin, E. (2015). NAD+ in aging, metabolism, and neurodegeneration. Science 350(6265): 1208-1213.
- Yang, H., Yang, T., Baur, J. A., Perez, E., Matsui, T., Carmona, J. J., Lamming, D. W., Souza-Pinto, N. C., Bohr, V. A., Rosenzweig, A., de Cabo, R., Sauve, A. A. and Sinclair, D. A. (2007). Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130(6): 1095-1107.
- Yoshino, J., Baur, J. A. and Imai, S. I. (2018). NAD+ intermediates: The biology and therapeutic potential of NMN and NR. Cell Metab 27(3): 513-528.
- Yoshino, J., Mills, K. F., Yoon, M. J. and Imai, S. (2011). Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14(4): 528-536.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kanamori, K. S., de Oliveira, G. C., Auxiliadora-Martins, M., Schoon, R. A., Reid, J. M. and Chini, E. N. (2018). Two Different Methods of Quantification of Oxidized Nicotinamide Adenine Dinucleotide (NAD+) and Reduced Nicotinamide Adenine Dinucleotide (NADH) Intracellular Levels: Enzymatic Coupled Cycling Assay and Ultra-performance Liquid Chromatography (UPLC)-Mass Spectrometry. Bio-protocol 8(14): e2937. DOI: 10.21769/BioProtoc.2937.
Category
Biochemistry > Other compound > NAD+/NADH
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












