- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Characterization of Protein Domain Function via in vitro DNA Shuffling
Published: Vol 8, Iss 11, Jun 5, 2018 DOI: 10.21769/BioProtoc.2873 Views: 6636
Reviewed by: Modesto Redrejo-RodriguezElizabeth LibbyChijioke Joshua

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ARP2/3 Phosphorylation Assay in the Presence of Recombinant Bacterial Effectors
Céline Michard [...] Patricia Doublet
Apr 5, 2017 9165 Views

In vitro and in vivo Assessment of Protein Acetylation Status in Mycobacteria
Krishna K. Singh [...] Deepak K. Saini
Jul 5, 2019 5666 Views

In vitro Glutamylation Inhibition of Ubiquitin Modification and Phosphoribosyl-Ubiquitin Ligation Mediated by Legionella pneumophila Effectors
Alan G. Sulpizio [...] Yuxin Mao
Nov 5, 2020 3921 Views
Abstract
We recently investigated the molecular events that drive evolution of the CTX-M-type β-lactamases by DNA shuffling of fragments of the blaCTX-M-14 and blaCTX-M-15 genes. Analysis of a total of 51 hybrid enzymes showed that enzymatic activity could be maintained in most cases, yet the enzymatically active hybrids were found to possess much fewer amino acid substitutions than the few hybrids that became inactive, suggesting that point mutations in the constructs rather than reshuffling of the fragments of the two target genes would more likely cause disruption of CTX-M activity. Certain important residues that played important functional roles in mediating enzyme activity were identified. These findings suggest that DNA shuffling is an effective approach to identify and characterize important functional domains in bacterial proteins.
Keywords: CTX-M-14Background
DNA recombination is a natural process by which genetic materials are exchanged among bacteria to enhance survival fitness under environmental stresses. Several hybrid CTX-M-lactamases (CTX-M-64, CTX-M-123, CTX-M-137, and CTX-M-132), presumably resulting from recombination between the blaCTX-M-14 and blaCTX-M-15 genes, the most common variants worldwide, have been reported in recent years (Nagano et al., 2009; Tian et al., 2014; He et al., 2015; Liu et al., 2015). Among these hybrid enzymes, CTX-M-64, which contained the N- and C-terminal portions of CTX-M-15 and the middle fragment of CTX-M-14, exhibited even higher catalytic activity than their parental prototypes (He et al., 2015).
DNA shuffling is a molecular approach designed to mimic and accelerate the evolution process through PCR-mediated random combinations of two target genes (Crameri et al., 1998). Our previous study demonstrated the use of DNA shuffling to investigate the molecular events driving the evolution of the CTX-M-type β-lactamases (Po et al., 2017). Mutants with or without cefotaximase activity were recovered. Important amino acid residues that played a role in conferring enzyme activity were identified by comparative analysis of the genotypes and phenotypes of the mutants. Such approach can be employed to characterize other functional proteins. Here we describe a detailed protocol of in vitro DNA shuffling.
Materials and Reagents
- 96-well cell culture plate (SPL Life Sciences, catalog number: 30096 )
- Test tubes
- Cotton swab (HUBY-340-CA-006)
- Filter (Pall, catalog number: 4612 )
- Petri dish (Corning, GosselinTM, catalog number: SB93-101 )
- E. coli DH5α (Thermo Fisher Scientific, InvitrogenTM, catalog number: 12297016 )
- E. coli BL21 (Thermo Fisher Scientific, InvitrogenTM, catalog number: C600003 )
- pET15b
- 5x Green GoTaq® Flexi Reaction Buffer (Promega, catalog number: M8911 )
- 2.5 mM dNTP mixture (Takara Bio, catalog number: 4030 )
- Magnesium chloride (UniChem, catalog number: M04550-4G )
- Primers (Synthesized by BGI)
- rTaq (Takara Bio, catalog number: R001WZ )
- PBS (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10010023 )
- 1x TAE (see Recipes)
- Milli-Q water
- QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- DNase I (Sigma-Aldrich, catalog number: DN25-1G )
- GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM1331 )
- Gel loading dye (New England Biolabs, catalog number: B7024S )
- SacI-HF (New England Biolabs, catalog number: R3156S )
- BamHI-HF (New England Biolabs, catalog number: R3136S )
- CutSmart® Buffer (New England Biolabs, catalog number: B7204S )
- T4 DNA Ligase (New England Biolabs, catalog number: M0202S )
- 10x T4 DNA Ligase Buffer (New England Biolabs, catalog number: B0202S )
- Tris (IBI Scientific, catalog number: IB70145 )
- EDTA (Merck, catalog number: 1.08421.1000 )
- Glacial acetic acid (DUKSAN, catalog number: 3839 )
- Agarose, Molecular Biology Certified (IBI Scientific, catalog number: IB70045 )
- 10,000x Gold View I (West Gene, catalog number: WGO-1 )
- LB Broth (Hopebio, catalog number: HB0128 )
- LB Nutrient Agar (Hopebio, catalog number: HB0129 )
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A9518-100G )
- QIAprep Spin Miniprep Kit (QIAGEN, catalog number: 27106 )
- Isopropyl-β-D-thiogalactopyranoside, IPTG (Santa Cruz Biotechnology, catalog number: sc-202185B )
- Mueller-Hinton broth, MH broth (BD, BBLTM, catalog number: 212322 )
- Mueller-Hinton Agar, MH agar (Hopebio, catalog number: HB6232 )
- Cefotaxime sodium salt (Sigma-Aldrich, catalog number: C7912-1G )
- Sodium chloride (VWR, catalog number: VWRC27810.295 )
- 50 mM Magnesium chloride (see Recipes)
- 50x TAE buffer (see Recipes)
- 1% Agarose gel (see Recipes)
- 10x DNase I reaction mixture (see Recipes)
- LB broth (see Recipes)
- 25 mg/ml Ampicillin (see Recipes)
- 0.5 M IPTG (see Recipes)
- MH broth (see Recipes)
- MH-IPTG (see Recipes)
- MH agar (see Recipes)
- Saline (see Recipes)
Equipment
- Bio-Rad S1000TM thermal cycler (Bio-Rad Laboratories, model: S1000TM )
- Centrifuge (Hettich Instruments, model: Mikro 185 )
- Power pad for electrophoresis (Labnet International, model: ENDUROTM 300V , catalog number: E0303)
- Gel tank
- UV transilluminator
- Spectrophotometer (Hach, model: DR 2800TM )
- Conventional water bath (42 °C for transformation)
- Conventional autoclave
- Microwave
- pH meter
Software
- PyMOL software
- BioEdit Sequence Alignment Editor
Procedure
The outline of the protocol is shown in Figure 1.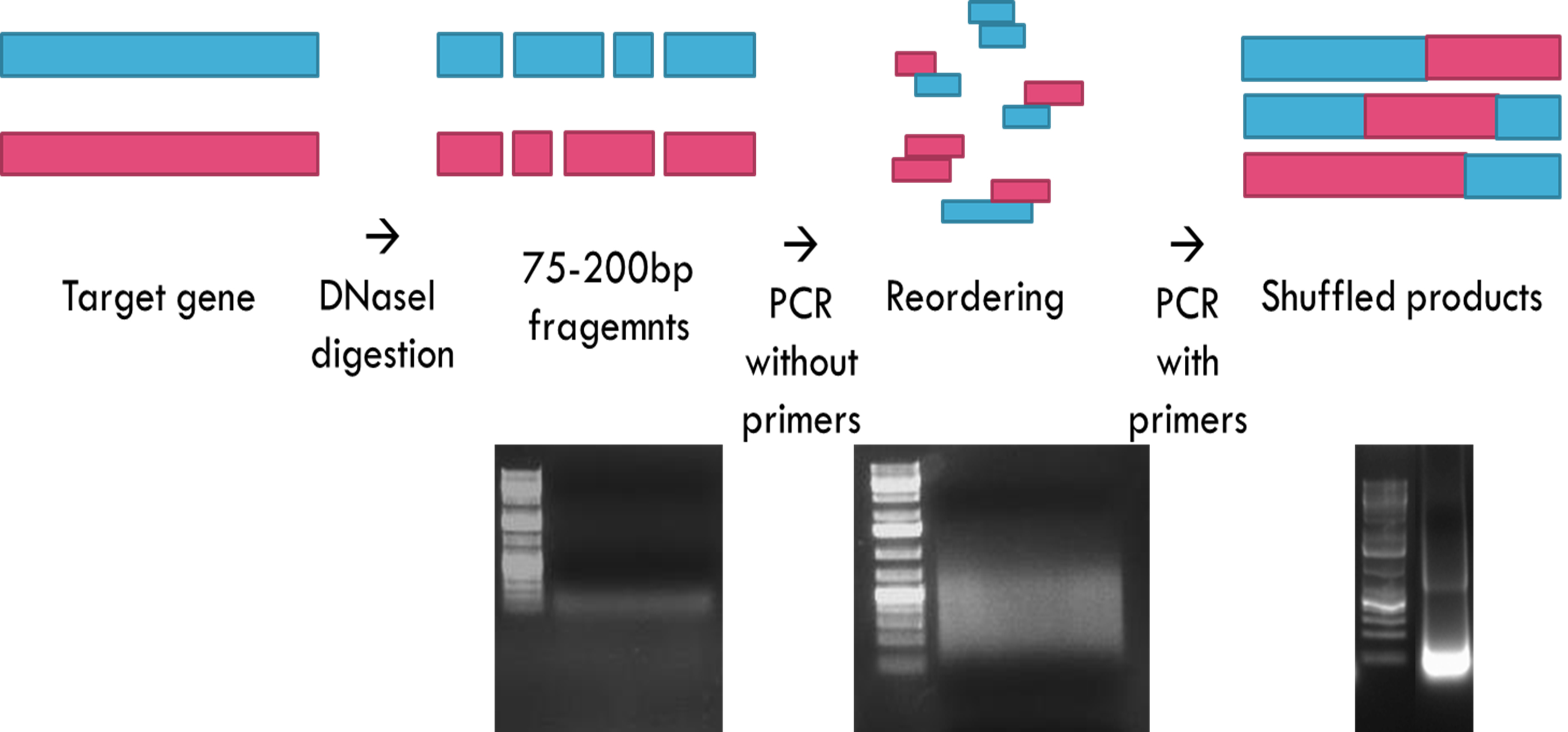
Figure 1. Scheme of DNA shuffling
- DNA shuffling
- Amplify the full length of target gene by PCR
Note: Forward and reverse primers should include a restriction site for subsequent cloning.- Prepare a 20 µl PCR reaction with 5x PCR buffer, 3 mM MgCl2, 200 μM deoxynucleoside triphosphates, 0.5 μM primers, and 1 U of Taq DNA polymerase.
- Add 1 µl DNA template.
Note: Prepare DNA templates by boiling 500 µl overnight culture in 300 µl PBS for 5 min. Collect the supernatant by centrifugation at 16,000 x g for 3 min. - Run PCR program (for target less than 1 kb):
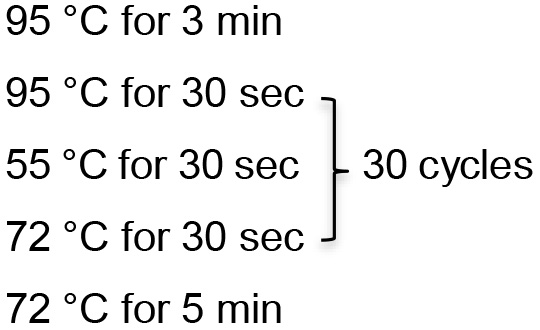
- Prepare a 20 µl PCR reaction with 5x PCR buffer, 3 mM MgCl2, 200 μM deoxynucleoside triphosphates, 0.5 μM primers, and 1 U of Taq DNA polymerase.
- Perform electrophoresis of PCR product in 1x TAE at 150 V for 15-30 min.
- Place the gel on a UV transilluminator (set at 365 nm wavelength).
- Cut DNA band and recover by gel extraction kit.
- Digest the 2 µg of DNA templates with 0.02 unit of DNase I and 10x DNase I reaction mixture in 100 µl reaction at room temperature for 10-20 min.
- Add gel loading dye.
- Run gel electrophoresis and recover DNA fragments of 50 to 200 bp by gel extraction kit.
- Reassemble the fragments by PCR without primers.
- Prepare a 100 µl PCR reaction with 5x PCR buffer, 3 mM MgCl2, 200 μM deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase.
- Use 10 - 30 ng/μl of the purified fragments as template.
- Run the program:
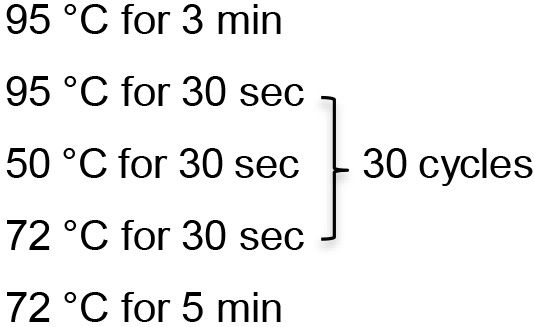
- Prepare a 100 µl PCR reaction with 5x PCR buffer, 3 mM MgCl2, 200 μM deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase.
- Purify PCR products of the correct size (depending on the size of the target gene) by gel extraction kit upon electrophoresis.
- Perform PCR again under conditions described above, except that 0.5 μM of each primer is used this time.
Note: The protocol worked when a DNA band with a size similar to that of the target genes was observed. If the concentration of recovered DNA was low, a larger amount could be added in subsequent restriction digestion reaction.
- Amplify the full length of target gene by PCR
- Cloning of target gene into expression vector
- Digest 600 ng of PCR products and pET15b separately with 0.6 µl SacI-HF, 0.6 µl BamHI-HF and 10x CutSmart® Buffer in 20 µl reaction.
Note: Include a restriction site in the forward and reverse primers if the restriction sites chosen were not present in the wild type target gene. However, the generation of restriction site was not guaranteed on reshuffled mutants. - Perform gel electrophoresis and recover DNA by gel extraction kit.
- Ligate with 5-10 ng DNA, 20-30 ng vector, 1 µl T4 DNA Ligase and 10x T4 ligase buffer in 20 µl reaction at 16 °C overnight.
- Transform the vector into E. coli DH5α.
Perform the transformation following the protocol provided by the manufacturer with modifications. See below:- Incubate 10 µl vector with 100 µl competent cell on ice for 30 min.
- Heat shock for 2 min.
- Incubate on ice for 5 min.
- Add 500 µl LB broth.
- Incubate at 37 °C for 1 h.
- Select on 100 µg/ml ampicillin plates.
- Incubate 10 µl vector with 100 µl competent cell on ice for 30 min.
- Perform colony PCR using the same primers and condition in A1 to check the presence of insert.
- Perform sequencing on transformants which contain an insert.
- Extract plasmids by QIAprep Spin Miniprep Kit.
- Transform E. coli BL21 competent cell with plasmids containing a mutant gene.
- Digest 600 ng of PCR products and pET15b separately with 0.6 µl SacI-HF, 0.6 µl BamHI-HF and 10x CutSmart® Buffer in 20 µl reaction.
- Antimicrobial susceptibility tests
- Add 150 µl MH-IPTG into each well of a 96 well plate.
- Add another 150 µl MH-IPTG broth into the last column of the 96 well plate.
- Add antibiotic into the last column. The test range depends on resistance breakpoints of different antibiotics; 512 µg/ml is the maximum concentration used for cefotaxime.
- Perform two-fold serial dilution along the row.
- Transfer 3 ml saline into n (number of test strains) test tubes.
- Transfer overnight culture of test strains from MH agar to test tubes prepared in Step C5 by cotton swab to obtain a suspension with OD600 0.08-0.1.
- Transfer 5 µl of suspension into each well of the 96 well plate.
- Incubate at 37 °C overnight.
- Record the minimum inhibition concentration (MIC).
Note: MIC is the minimal test concentration at which no bacterial growth was observed.
- Add 150 µl MH-IPTG into each well of a 96 well plate.
Data analysis
Sequence the target insert in two directions (forward and reverse). Align the sequences of mutants with that of the wildtype to identify mutations. Obtain protein structures from the Protein Data Bank (http://www.rcsb.org/pdb/). Analyze protein structures with the PyMOL software.
- Analysis of the sequencing data by BioEdit (Figure 2)
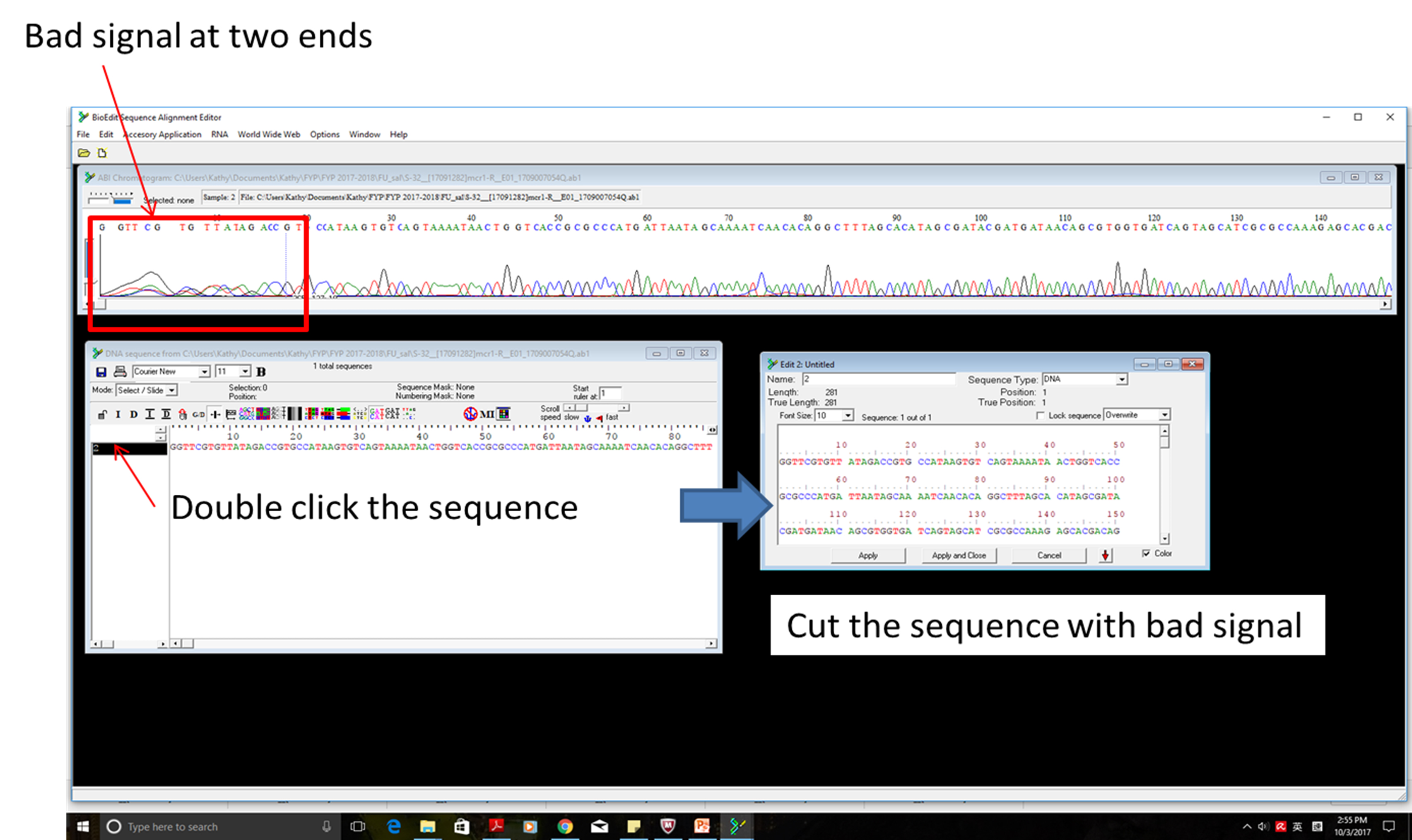
Figure 2. Analysis of sequencing data- Edit→Copy Sequences to the clipboard (Fasta Format)
- Import the reverse sequence together with the forward sequence: File→Import from Clipboard
- Join the forward and reverse sequence together: Highlight both sequences→click ‘Accessory Application’→CAP contig assembly program
- Align the sequence with wild type (Figure 3)
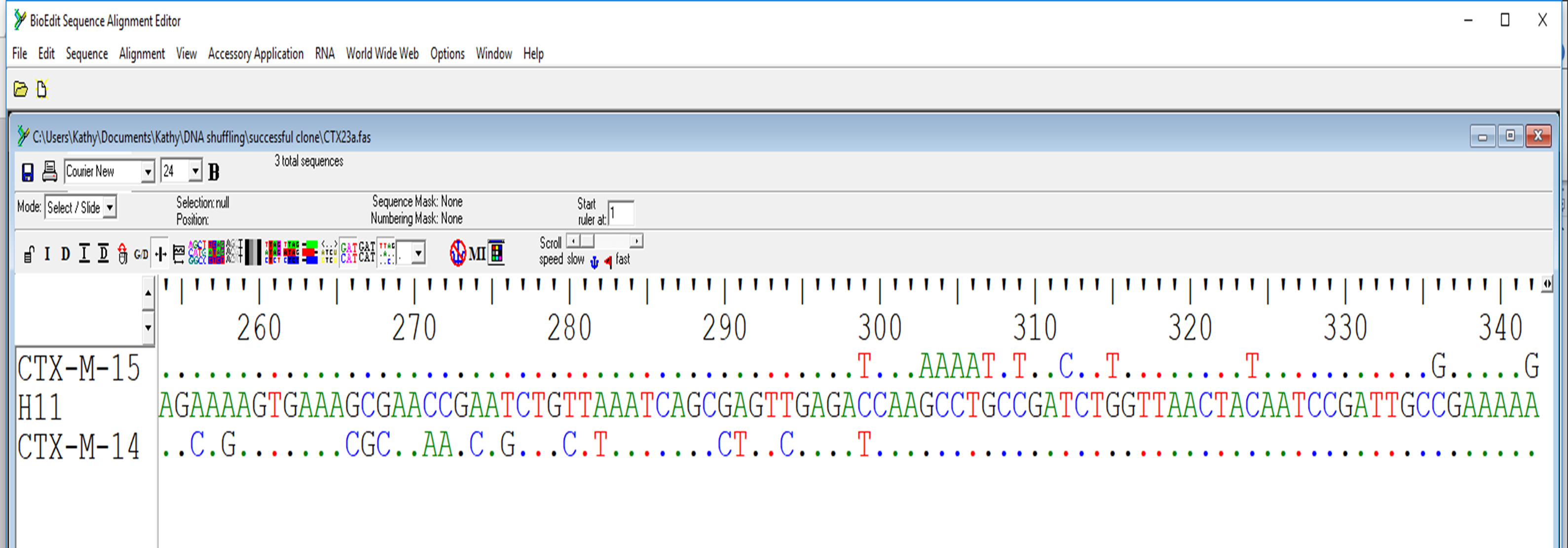
Figure 3. Alignment of mutant with wild type sequence. H11 was generated by recombination of CTX-M-14 and CTX-M-15. Sequence before nucleotide number 299 was from CTX-M-15 while that after 299 was from CTX-M-14. - Edit→Copy Sequences to the clipboard (Fasta Format)
- Analysis of protein structure by PyMOL
Proteins in which specific amino acid substitutions caused a significant reduction in MIC were subjected to structural analysis. Locations of the amino acids concerned were visualized by PyMOL. Observation of local environment and identification of possible interaction between nearby residues helped to figure out the molecular basis of phenotypic changes. For the detailed structure information of CTX-M-14 by PyMOL, please see Po et al., 2017.
Recipes
- 50 mM Magnesium chloride
Dissolve 0.238 g MgCl2 into 50 ml ddH2O
Autoclave at 121 °C for 15 min - 50x TAE buffer
Dissolve 242 g Tris into 600 ml ddH2O
Add 57.1 ml glacial acetic acid and 100 ml 0.5 M EDTA
Adjust the pH to 8
Add ddH2O to 1 L
Final (1x) working concentration:
0.04 M Tris
0.001 M EDTA - 1x TAE buffer
Mix 100 ml 50x TAE buffer will 4,900 ml ddH2O - 1% Agarose gel
Mix 1 g agarose with 100 ml 1x TAE buffer
Heat to dissolve by microwave
Add 1 µl 10,000x Gold View I - 10x DNase I reaction mixture
Dissolve:
6 g Tris
0.095 g MgCl2
Adjust pH to 7.4
Add ddH2O to 100 ml - LB broth
Dissolve 12.5 g of LB powder into 500 ml ddH2O
Autoclave at 121 °C for 15 min - 25 mg/ml Ampicillin
Dissolve 1.25 g ampicillin sodium salt into 50 ml ddH2O
Filter sterilize - 100 µg/ml Ampicillin plate
Dissolve 20 g of LB powder into 500 ml ddH2O
Autoclave at 121 °C for 15 min
Add 2 ml 25 mg/ml ampicillin - 0.5 M IPTG
Dissolve 5.96 g IPTG into 50 ml ddH2O
Filter sterilize - MH broth
Dissolve 11 g of MH broth powder into 500 ml ddH2O
Autoclave at 121 °C for 15 min - MH-IPTG
Add 100 µl 0.5 M IPTG into 50 ml MH broth - MH agar
Dissolve 21 g of MH agar powder into 500 ml ddH2O
Autoclave at 121 °C for 15 min - Saline
Dissolve 4.5 g sodium chloride into 500 ml ddH2O
Autoclave at 121 °C for 15 min
Acknowledgments
This work has been previously published in Po et al., 2017. This work was supported by the Collaborative Research Fund of the Research Grant Council (C7038-15G and C5026-16G), and the Health and Medical Research Fund of the Food and Health Bureau, the Government of the Hong Kong SAR (HMRF: 14130422 to SC).
Competing interests
Potential conflicts of interest: All authors declare there are no conflicts to disclose. All authors declare there are no conflicts to disclose.
References
- Crameri, A., Raillard, S. A., Bermudez, E. and Stemmer, W. P. (1998). DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391(6664): 288-291.
- He, D., Chiou, J., Zeng, Z., Liu, L., Chen, X., Zeng, L., Chan, E. W., Liu, J. H. and Chen, S. (2015). Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob Agents Chemother 59(10): 5976-5983.
- Liu, L., He, D., Lv, L., Liu, W., Chen, X., Zeng, Z., Partridge, S. R. and Liu, J. H. (2015). blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrob Agents Chemother 59(8): 4464-4470.
- Nagano, Y., Nagano, N., Wachino, J., Ishikawa, K. and Arakawa, Y. (2009). Novel chimeric β-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 β-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob Agents Chemother 53(1): 69-74.
- Po, K. H. L., Chan, E. W. C. and Chen, S. (2017). Functional characterization of CTX-M-14 and CTX-M-15 β-lactamases by in vitro DNA Shuffling. Antimicrob Agents Chemother 61(12).
- Tian, G. B., Huang, Y. M., Fang, Z. L., Qing, Y., Zhang, X. F. and Huang, X. (2014). CTX-M-137, a hybrid of CTX-M-14-like and CTX-M-15-like β-lactamases identified in an Escherichia coli clinical isolate. J Antimicrob Chemother 69(8): 2081-2085.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Po, K. H. L., Chan, E. W. C. and Chen, S. (2018). Characterization of Protein Domain Function via in vitro DNA Shuffling. Bio-protocol 8(11): e2873. DOI: 10.21769/BioProtoc.2873.
Category
Microbiology > Microbial biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








