- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Hyaluronan Isolation from Mouse Mammary Gland
Published: Vol 8, Iss 11, Jun 5, 2018 DOI: 10.21769/BioProtoc.2865 Views: 6089
Reviewed by: Vivien Jane Coulson-Thomastarsis ferreiraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Technique for the Measurement of in vitro Phospholipid Synthesis via Radioactive Labeling
Lucia Rodriguez-Berdini and Gabriel O. Ferrero
Jan 20, 2016 14628 Views

Liposome Flotation Assays for Phosphoinositide-protein Interaction
Helene Tronchere and Frederic Boal
Mar 5, 2017 19196 Views

Enhanced Ribonucleoprotein Immunoprecipitation (RIP) Technique for the Identification of mRNA Species in Ribonucleoprotein Complexes
Saja A. Fakhraldeen [...] Caroline M. Alexander
Oct 5, 2022 2855 Views
Abstract
The glycosaminoglycan hyaluronan (HA) is a key component of the extracellular matrix. The molecular weight of HA is heterogeneous and can reach from several million to several hundred daltons. The effect of HA on cell behavior is size dependent; fragmented HA acts as a danger signal, stimulates cell migration and proliferation and is proinflammatory, native high molecular weight HA suppresses inflammation. Therefore, it is important to analyze HA size distribution when studying the role of HA in tissue homeostasis and pathology. This protocol describes isolation of HA from mouse mammary glands but can also be applied to other tissues. The quality of the isolated HA is sufficient to analyze size distribution by gel electrophoresis (Calabro et al., 2000).
Keywords: HyaluronanBackground
The glycosaminoglycan HA consists of N-acetyl glucosamine and beta glucuronic acid disaccharides and is a ubiquitous component of the extracellular matrix. High molecular weight HA is fragmented by enzymatic degradation and oxidation by reactive oxygen and nitrogen species. In healthy tissues, the majority of HA is of high molecular weight. Accumulation of fragmented HA acts as danger signal during pathological processes (Tolg et al., 2012 and 2017; Yuan et al., 2015). For example, HA fragments stimulate inflammation whereas high molecular weight HA suppresses inflammation. HA influences cell behavior such as cell migration and proliferation by interaction with cell membrane receptors, leading to activation of signaling pathways. Since receptor-HA interactions are influenced by HA size, the effect of HA on tissue biology depends not only on HA amount but rather on HA size distribution and HA receptor expression by individual cells.
Materials and Reagents
- Pipette tips (Fisher Scientific, catalog numbers: 02-707-417 , 02-707-408 , 02-707-438 )
- 1.5 ml Eppendorf microcentrifuge tubes (Diamed Advan Tech, catalog number: AD150-N )
- Slide-A-Lyzer® MINI Dialysis Devices 7K MWCO, 0.1 ml (Thermo Fisher Scientific, catalog number: 69560 )
- Slide-A-Lyzer® MINI Dialysis Devices 3.5K MWCO, 0.5 ml/2 ml (Thermo Fisher Scientific, catalog number: 88400 / 88403 )
- Pierce Strong Anion Exchange Spin Column, Mini (Thermo Fisher Scientific, catalog number: 90010 )
- C57Bl/6 female mice
- Sterile de-ionized water
- NaOH (BioShop, catalog number: SHY700.500 )
- Tris base (121.1 g/mole) (BioShop, catalog number: TRS001.1 )
- NaCl (58.44 g/mole) (BioShop, catalog number: SOD001.10 )
- SDS (BioShop, catalog number: SDS001 )
- CaCl2·2H2O (147.0 g/mole) (Sigma-Aldrich, catalog number: C3306 )
- PBS (WISENT, catalog number: 311-425-CL )
- Proteinase K (Roche Diagnostics, catalog number: 03115879001 ), recombinant, PCR grade
Note: It is approximately 2.5 U/mg when assayed with the Chromozym assay, or 30 U/mg with the hemoglobin assay. - Chloroform (reagent grade, Acros Organics, catalog number: 423555000 )
- Ethanol (EtOH), 100%
- Deferoxamine mesylate salt (=deferoxamine methanesulfonate salt) (657 g/mole) (Sigma Aldrich, catalog number: 1166003 )
- Benzonase (Sigma-Aldrich, catalog number: E1014 ) recombinant 250 U/μl
- 0.2 M NaOH (see Recipes)
- 1 M Tris pH 8.3 (see Recipes)
- 5 M NaCl (see Recipes)
- 10% SDS (see Recipes)
- 1 M CaCl2 (see Recipes)
- Proteinase K solution, 20 mg/ml (see Recipes)
- Deferoxamine mesylate solution, 50 mg/ml (see Recipes)
- Tissue lysis buffer (see Recipes)
- 0.1 M NaCl (see Recipes)
- Benzonase digestion buffer (see Recipes)
- NaCl solutions for ion exchange (IEX) fractionation (see Recipes)
Equipment
- Pipettor (Drummond Scientific, Pipet-Aid®, model: XP2 )
- Pipetman (Gilson, model: PIPETMAN® Classic )
- Incubator capable to hold 37 °C (VWR, model: 1510 E )
- Eppendorf Thermomixer (Eppendorf, model: ThermoMixer® R )
- Refrigerated tabletop microcentrifuge capable of 18,000 x g (Thermo Fisher Scientific, model: SorvallTM LegendTM Micro 21R )
- Rotor included with SorvallTM LegendTM Micro 21R
- Flammable materials storage freezer, -20 °C
- Fisher Scientific stirring Hotplate
- Biohazard/Laminar flow hood
- Digital Analytical Balance Scale (U.S. Solid Store, model: USS-DBS5 ); Repeatability: ± 0.2 mg
- Balance scale (Denver Instrument, model: XS-310 D , Repeatability ± 0.02 g
- pH meter (Fisher Scientific, model: accumetTM AR15 )
- Fluorophore-assisted carbohydrate electrophoresis (FACE)
Procedure
Note: The dialysis and precipitation steps do not cause losses of HA fragments as small as 4 kDa.
- HA extraction from mouse mammary gland
Mammary glands were isolated from 8 weeks old C57Bl/6 female mice as described in Tolg et al. (in review) Steps 2-3. The typical HA yield is 5-15 µg/100 mg tissue.- Add 800 µl of tissue lysis buffer to 50-200 mg mammary gland tissue.
- Incubate samples 16-24 h at 55 °C. Mix samples during incubation, an Eppendorf Thermomixer at 500 rpm mixing speed works well.
- The following day, centrifuge digested samples at 18,000 x g (13,000 rpm) for 5 min at room temperature. This step removes undigested debris. Dispose of undigested debris as a biohazard.
- Pour the supernatant into a new Eppendorf microcentrifuge tube.
- Increase the NaCl concentration of the sample to 2 M by adding 11.7 mg NaCl powder/100 µl sample. Dissolve the NaCl powder by inverting samples end over end 10 times, then let samples sit for 1 min to allow undissolved NaCl crystals sink to the bottom of the tube where they will be visible. If necessary, repeat mixing until NaCl is dissolved. It takes a few minutes for the NaCl to dissolve.
- Remove proteinase K by chloroform extraction: Determine the sample volume and add the same volume of chloroform to the sample and mix by turning the sample upside down several times.
- Centrifuge the sample for 10 min at 18,000 x g (13,000 rpm) to separate aqueous and chloroform phases.
- Transfer the upper aqueous phase (Figure 1) into a new Eppendorf tube and repeat the chloroform extraction (Step A6).
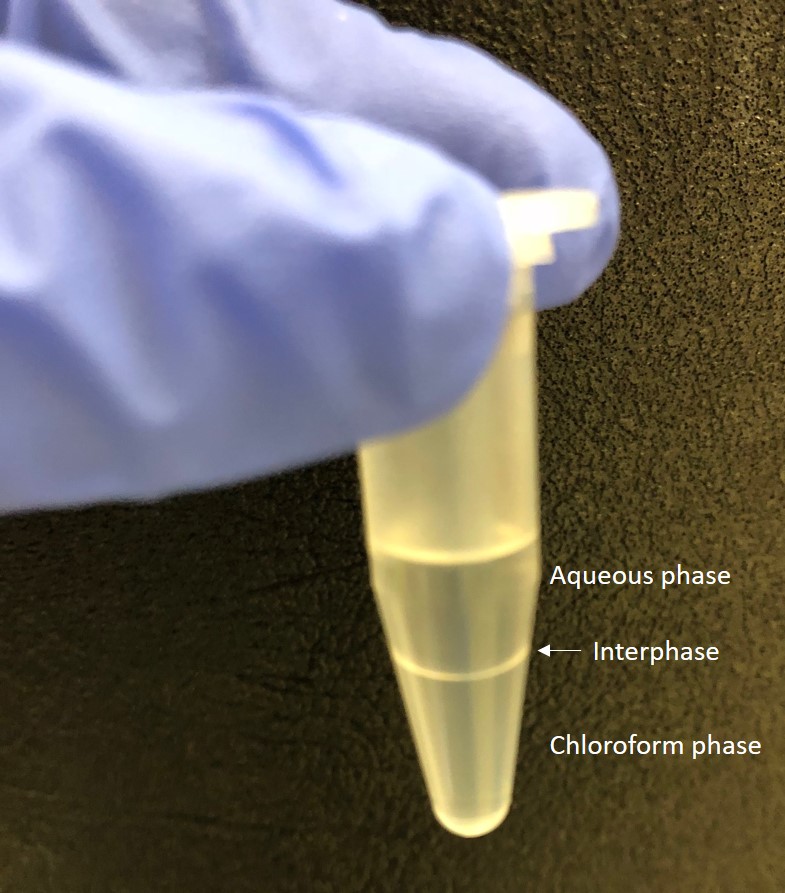
Figure 1. Separation of aqueous and chloroform phases. After centrifugation the aqueous (top) and chloroform (bottom) phases are clearly visible. - Centrifuge the sample for 10 min at 18,000 x g (13,000 rpm) and transfer the upper aqueous phase into a new Eppendorf tube.
- Reduce NaCl concentration by dialysis against 0.1 M NaCl: Remove 80% of the aqueous phase of each sample and transfer into two MINI Dialysis Units (7,000 MWCO, 0.1 ml capacity). Add the Dialysis Units to a beaker with 4 L of 0.1 M NaCl and dialyze for 2 h at 4 °C. During dialysis, slowly stir the 0.1 M NaCl using a magnetic stirring plate.
- Precipitate HA by Ethanol precipitation: Transfer samples from the Dialysis unit into 1.5 ml micro-centrifuge tubes, add 4 vol of 100% EtOH, mix and then store 16-24 h at -20 °C. Samples can be stored at -20 °C for up to one week.
- Centrifuge samples for 30 min at 18,000 x g (13,000 rpm), 4 °C.
- Remove supernatant and discard as chemical waste. Add 500 µl cold 70% EtOH to the pellet. Mix and repeat centrifugation for 10 min at 18,000 x g (13,000 rpm), 4 °C.
- Remove supernatant and discard as chemical waste. Allow HA pellet to dry by keeping sample tubes open under the cell culture or laminar flow hood for 1 h.
- Remove co-precipitated DNA by digestion with the nuclease Benzonase: Dissolve HA pellet in 100 µl of 50 mM Tris pH 8.3, 20 mM NaCl, containing 500 U Benzonase and digest for 4 h at 37 °C.
- Increase the NaCl concentration of the sample to 2 M by dissolving 11.7 mg NaCl powder directly in the sample as described in Step A5.
- Determine the sample volume and add the same volume of chloroform to the sample and mix by turning the sample upside down several times.
- Centrifuge the sample for 10 min at 18,000 x g (13,000 rpm) to separate aqueous and chloroform phases.
- Remove 80% of the aqueous phase of each sample and transfer into two MINI Dialysis Units 7,000 MWCO, 0.1 ml capacity. Add the Dialysis Units to a beaker with 4 L of 0.1 M NaCl and dialyze for 2 h at 4 °C. During dialysis, slowly stir the 0.1 M NaCl using a magnetic stirring plate.
- Transfer samples from the Dialysis unit, add 4 vol of 100% EtOH, mix then store 16-24 h at -20 °C. Samples can be stored at -20 °C for up to one week.
- Centrifuge samples for 30 min at 18,000 x g (13,000 rpm), 4 °C.
- Remove supernatant and add 500 µl cold 70% EtOH. Mix and repeat centrifugation for 10 min at 18,000 x g (13,000 rpm), 4 °C.
- Remove supernatant and discard as chemical waste. Allow HA pellet to dry by keeping sample vials open under the cell culture or laminar flow hood for 1 h.
- Dissolve HA in 50 µl deionized H2O.
- The yield is usually about 5-15 µg HA per 100 mg tissue.
- Add 800 µl of tissue lysis buffer to 50-200 mg mammary gland tissue.
- Further HA purification and size fractionation by ion exchange (IEX)
Notes:- This technique is a stepwise elution technique, involving pre-wash and equilibration of IEX spin columns, HA sample loading, and sample elution by applying NaCl solutions with increasing concentration. This technique is able to remove some non-HA contaminants and to fractionate a polydisperse HA sample into low polydispersity (i.e., narrow size range) fractions between ca. 7 and 100 kDa, but does not subfractionate the higher molecular mass HA components. No preferential sample loss for any size of HA in IEX fractionation is observed. Very high molecular mass HA can also be washed out from the column without sample loss, due to the porous structure (3µ) of the column material (a regenerated cellulose-based membrane).
- All relevant solutions and apparatus must be at room temperature before use. Temperature affects the size of HA eluted from the IEX column. If cold elution solutions are used, HA will be prematurely eluted at a lower salt concentration than expected. Elution reproducibility is also strongly dependent on centrifugation time and force applied to the IEX columns. The centrifugal force and time applied to the IEX spin column need to be kept constant, and the force is recommended to be no higher than 400 x g. Higher force can result in premature release of HA from the column.
- The passed liquid is collected in 1.5 ml microcentrifuge tubes with caps open and pointed toward the center of the rotor. Always align the printed letter Q in the IEX column toward the center of the fixed angle rotor, so that the solution can pass through the column resin uniformly. For each step, centrifuge at 400 x g for 2-5 min. It is recommended to spin for 2 min and check to see if the solution has passed. Spin for 1 min at a time and check repeatedly until all of the solution has passed. It is important not to over spin the columns. Do not wait for long periods between elution steps, as the column may dry out and prevent proper elution of the solutions and sample.
- If the sample volume is much larger than 400 µl, several columns may be used. This works better than repeated loading of a single column.
- The sample pH should be higher than ~5. The pKa for HA is ~3.2, and the pH of the NaCl solution is ~5.6.
- Elution using NaCl concentrations of 0.9 to 2.0 M elute sulfated glycosaminoglycans.
- Sample preparation
HA samples are loaded in 0.1 M NaCl. The HA stock concentration should be less than about 0.5 µg/µl. You will use 400 µl of each sample. The maximum load is about 200 µg per column. If starting with HA sample in de-ionized water, add 2 M NaCl to bring the final salt concentration to 0.1 M. - Pre-washing and equilibration of the IEX columns
We found that an unknown (possibly cationic) species may be shed from the columns during sample elution and later cause precipitation of HA following fractionation and dialysis. The IEX column is therefore pre-washed once with 400 µl of 0.1 M NaCl, then 3 times with 400 µl of 0.800 M NaCl, and then re-equilibrated using 5 washes with 400 µl of 0.100 M NaCl before use. - Sample loading on column
Load 400 µl HA sample solution (in 0.1 M NaCl) onto the column and centrifuge at 400 x g for 2-min. - Wash through unbound or weakly bound material
- Wash the column with 400 µl 0.1 M NaCl to fully elute unbound neutral or positively charged contaminants.
- Then wash the column with 400 µl 0.2 M NaCl three times to remove any weakly bound impurities. Only HA less than ~4 kDa (10 disaccharides long) will elute at this concentration. Fractions collected using 0.2 M NaCl can be analyzed for HA oligomers by fluorophore-assisted carbohydrate electrophoresis (FACE).
- Wash the column with 400 µl 0.1 M NaCl to fully elute unbound neutral or positively charged contaminants.
- HA Fraction Elution
Elute HA from low to high molecular mass by increasing NaCl concentration in each elution. For each elution, 2 x washes are needed to completely wash out that HA fraction. Combine the elutions from the same NaCl concentrations for dialysis. HA will be eluted out by NaCl solutions from 0.3 to 0.8 M. The molecular mass of HA eluted at a range of NaCl concentrations are listed below (Table 1 and Table 2).
Table 1. Summary IEX Steps, 4 step method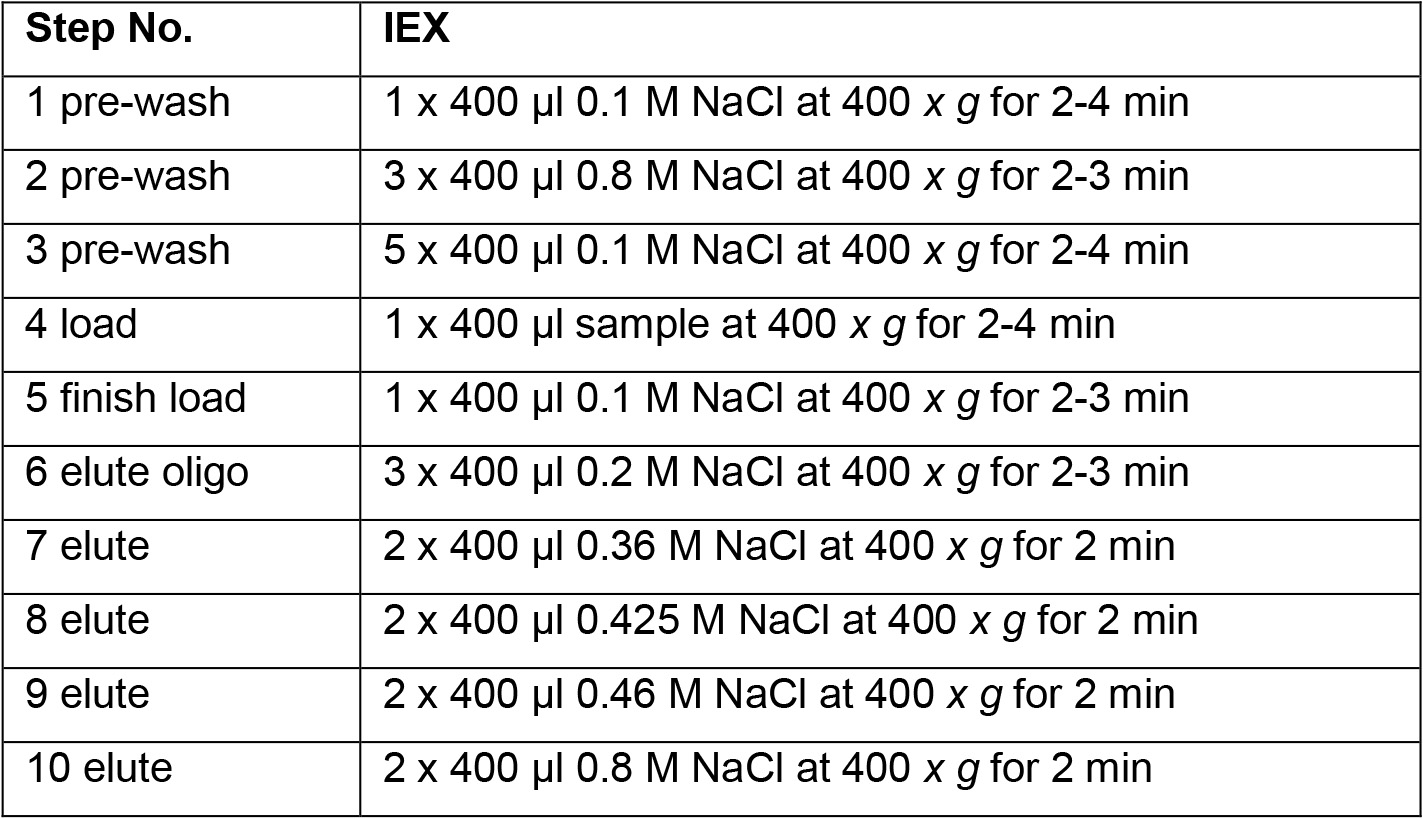
Table 2. HA size characteristics eluted at different NaCl concentrations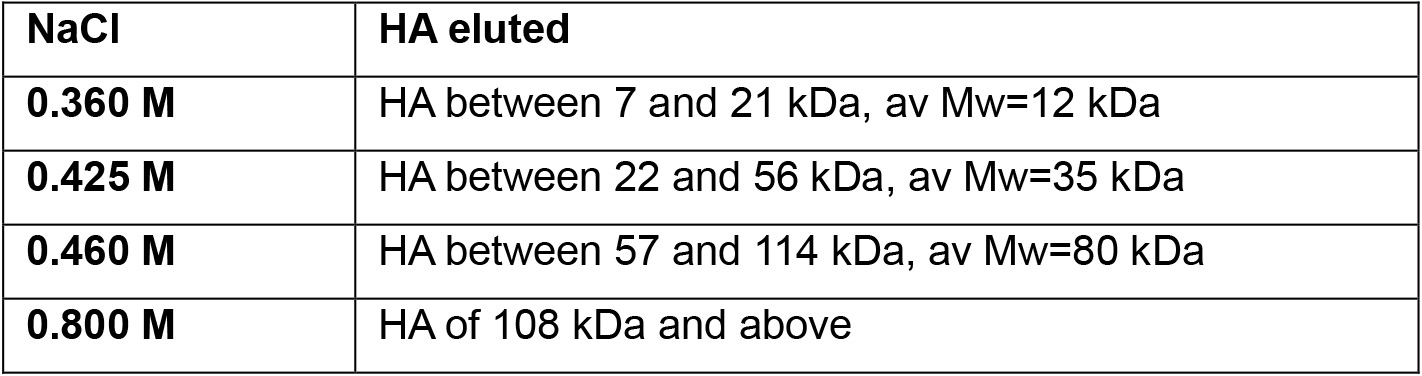
- Removal of NaCl from HA fractions: Dialyze exhaustively to de-ionized water before concentrating or drying. Centrifugal concentration of HA solutions containing salt can cause HA degradation. Alternatively, precipitate HA with 4 vol EtOH.
- This technique is a stepwise elution technique, involving pre-wash and equilibration of IEX spin columns, HA sample loading, and sample elution by applying NaCl solutions with increasing concentration. This technique is able to remove some non-HA contaminants and to fractionate a polydisperse HA sample into low polydispersity (i.e., narrow size range) fractions between ca. 7 and 100 kDa, but does not subfractionate the higher molecular mass HA components. No preferential sample loss for any size of HA in IEX fractionation is observed. Very high molecular mass HA can also be washed out from the column without sample loss, due to the porous structure (3µ) of the column material (a regenerated cellulose-based membrane).
Data analysis
The purified HA fractions can be quantified using an ELISA kit (for example kit K-1200, ECHELON) or by agarose gel electrophoresis and image analysis.
Recipes
- 0.2 M NaOH
Dissolve 0.8 g NaOH in 100 ml deionized water - 1 M Tris pH 8.3
- Dissolve 12.1 g Tris base powder in 80 ml de-ionized water
- Adjust pH to 8.3 with 12 N HCl
- Add deionized water to final volume of 100 ml
- Autoclave
- Dissolve 12.1 g Tris base powder in 80 ml de-ionized water
- 5 M NaCl
- Dissolve 29.22 g NaCl in deionized water to final volume of 100 ml
- Autoclave
- Dissolve 29.22 g NaCl in deionized water to final volume of 100 ml
- 10% SDS
Dissolve 10 g SDS powder in 100 ml deionized water - 1 M CaCl2
- Dissolve 14.7 g CaCl2·2H2O in deionized water to final volume of 100 ml
- Autoclave
- Dissolve 14.7 g CaCl2·2H2O in deionized water to final volume of 100 ml
- Proteinase K solution, 20 mg/ml
Note: Ca. 25 U/ml when assayed by the Chromozym assay, or 300 U/ml using the hemoglobin assay.- Dissolve 20 mg Proteinase K in 1 ml PBS buffer
- Divide into 0.1 ml aliquots and store at -20 °C
- Dissolve 20 mg Proteinase K in 1 ml PBS buffer
- Deferoxamine mesylate solution, 50 mg/ml (= 0.076 M)
- Dissolve 50 mg deferoxamine mesylate in 1 ml deionized water
- Divide into 50 μl aliquots and store at -20 °C
- Dissolve 50 mg deferoxamine mesylate in 1 ml deionized water
- Tissue lysis buffer
- Premix buffer stock:
15 ml 1 M Tris pH 8.3
3 ml 5 M NaCl
1 ml 1 M CaCl2
1 ml 10% SDS
80 ml sterile deionized water
Note: This buffer stock composition is 0.15 M Tris pH 8.3, 0.15 M NaCl, 0.01 M CaCl2, 0.1% SDS. - Just before use, add 100 µl Proteinase K solution and 657 µl deferoxamine mesylate solution to 10 ml above buffer stock.
- Final composition in tissue lysis buffer is:
0.2 mg/ml Proteinase K (2.5 U/ml by Chromzym assay, or 30 U/ml by hemoglobin assay)
3.285 mg/ml (0.005 M) deferoxamine mesylate
- Premix buffer stock:
- 0.1 M NaCl
Dissolve 5.8 g NaCl in deionized water to final volume of 1 L - Benzonase digestion buffer
50 mM Tris pH 8.3,
20 mM NaCl
500 U/ml Benzonase- Add 5 ml 1 M Tris pH 8.3 and 0.4 ml 5 M NaCl to 94.6 ml deionized sterile water
- Just before use, add 20 µl Benzonase to 1 ml digestion buffer
- Add 5 ml 1 M Tris pH 8.3 and 0.4 ml 5 M NaCl to 94.6 ml deionized sterile water
- NaCl solutions for ion exchange (IEX) fractionation
Note: Prepare the NaCl solutions one day prior to IEX experiment. Leave all solutions at room temperature overnight.- 2.00 M NaCl stock solution for IEX: Dissolve 11.688 g NaCl in deionized
- water to 100 ml final volume
- Diluted NaCl solutions for washing and elution of IEX columns
- Make dilutions of the 2 M NaCl stock solution as described below:
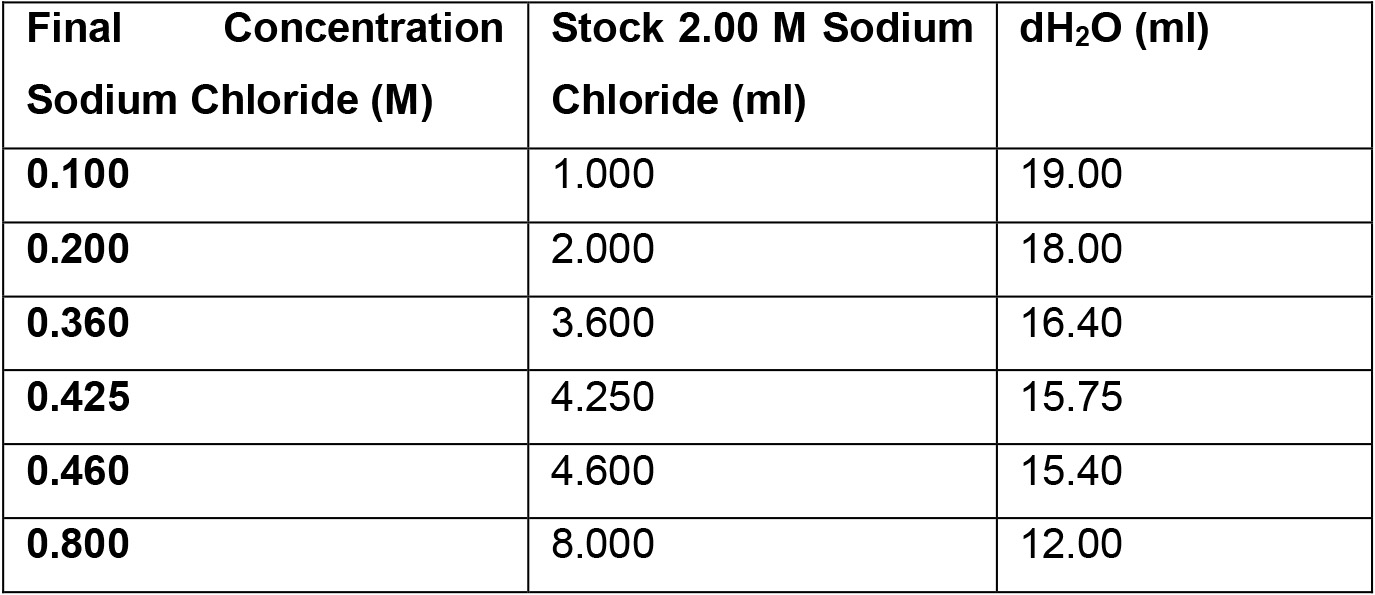
- 2.00 M NaCl stock solution for IEX: Dissolve 11.688 g NaCl in deionized
Acknowledgments
This work was supported by a CBCF grant to ET and The Endre A. Balazs Foundation to MC. This protocol was adapted from Tolg et al. (2012 and 2017) and Yuan et al. (2015).
Competing interests
Authors have no conflict of interest.
References
- Calabro, A., Benavides, M., Tammi, M., Hascall, V. C. and Midura, R. J. (2000). Microanalysis of enzyme digests of hyaluronan and chondroitin/dermatan sulfate by fluorophore-assisted carbohydrate electrophoresis (FACE). Glycobiology 10(3): 273-281.
- Tolg, C., Cowman, M., and Turley E. A. (2018). Mouse mammary gland whole mount preparation and analysis. Bio-protocol 8(13): e2915.
- Tolg, C., Hamilton, S. R., Zalinska, E., McCulloch, L., Amin, R., Akentieva, N., Winnik, F., Savani, R., Bagli, D. J., Luyt, L. G., Cowman, M. K., McCarthy, J. B. and Turley, E. A. (2012). A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am J Pathol 181(4): 1250-1270.
- Tolg, C., Yuan, H., Flynn, S. M., Basu, K., Ma, J., Tse, K. C. K., Kowalska, B., Vulkanesku, D., Cowman, M. K., McCarthy, J. B. and Turley, E. A. (2017). Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biol 63: 117-132.
- Yuan, H., Amin, R., Ye, X., de la Motte, C. A. and Cowman, M. K. (2015). Determination of hyaluronan molecular mass distribution in human breast milk. Anal Biochem 474: 78-88.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tolg, C., Cowman, M. and Turley, E. A. (2018). Hyaluronan Isolation from Mouse Mammary Gland. Bio-protocol 8(11): e2865. DOI: 10.21769/BioProtoc.2865.
Category
Cancer Biology > General technique > Biochemical assays > Other compound
Developmental Biology > Morphogenesis > Organogenesis
Cell Biology > Tissue analysis > Physiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








