- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vivo Use of Dextran-based Anterograde Cortical Tracers to Assess the Integrity of the Cortical Spinal Tract
Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2862 Views: 7647
Reviewed by: Oneil G. BhalalaTrevor Martin SmithEhsan Kheradpezhouh

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Conditioned Lick Suppression: Assessing Contextual, Cued, and Context-cue Compound Fear Responses Independently of Locomotor Activity in Mice
Youcef Bouchekioua [...] Yu Ohmura
Dec 5, 2022 1641 Views

In situ Microinflammation Detection Using Gold Nanoclusters and a Tissue-clearing Method
Fayrouz Naim [...] Masaaki Murakami
Apr 5, 2023 2670 Views

A One-Step Mouse Model of Parkinson’s Disease Combining rAAV-α-Synuclein and Preformed Fibrils of α-Synuclein
Santhosh Kumar Subramanya [...] Poonam Thakur
Dec 5, 2025 1644 Views
Abstract
When injected into the motor cortex of rats, anterograde tracers label fibers of the associated descending corticospinal tract (CST) that originate from pyramidal neurons in the tracer-injected cortex. These fibers can be assessed at the level of the spinal cord to determine the integrity of the descending CST and the spatial distribution of axons in the spinal grey matter. Here we provide detailed methods on the minimally invasive stereotaxic injection of anterograde tracers into the forelimb sensorimotor representation in the rat cortex. In addition, we detail the fixing and processing of spinal tissue for assessment of CST integrity and branching into spinal grey matter.
Keywords: Cortical spinal tractBackground
The motor cortex contains upper motor neurons which give rise to descending axons that form the CST and terminate in the spinal cord. Anterograde tracers injected into the motor cortex of rats label these descending fibers of the CST allowing their terminals to be assessed at the level of the spinal cord (Figure 1). In rodent models, anterograde labeling of the CST is widely used to assess CST integrity in a range of spinal cord injury models including lesions (Thallmair et al., 1998; Vavrek et al., 2006) and contusions (Hill et al., 2001). Further, anterograde cortical tracers have been used to assess spared fibers and plasticity of the CST in various models of brain injury such as cortical stroke (Wahl et al., 2014; Wiersma et al., 2017), traumatic brain injury (Ueno et al., 2012) and pyramidotomy (Starkey et al., 2012). Here we provide detail methods on injection of anterograde tracers into motor cortex corresponding the forelimb. We describe how to create burr holes without inducing damage to the cortex and the use of pulled glass pipette tips for minimally invasive injection. Since the CST is widely assessed in both brain and spinal cord injury models, we offer a systematic method of assessing the CST and branching fibers in the surrounding spinal grey matter that accounts for variability due to inter-animal differences in tracer efficacy or uptake.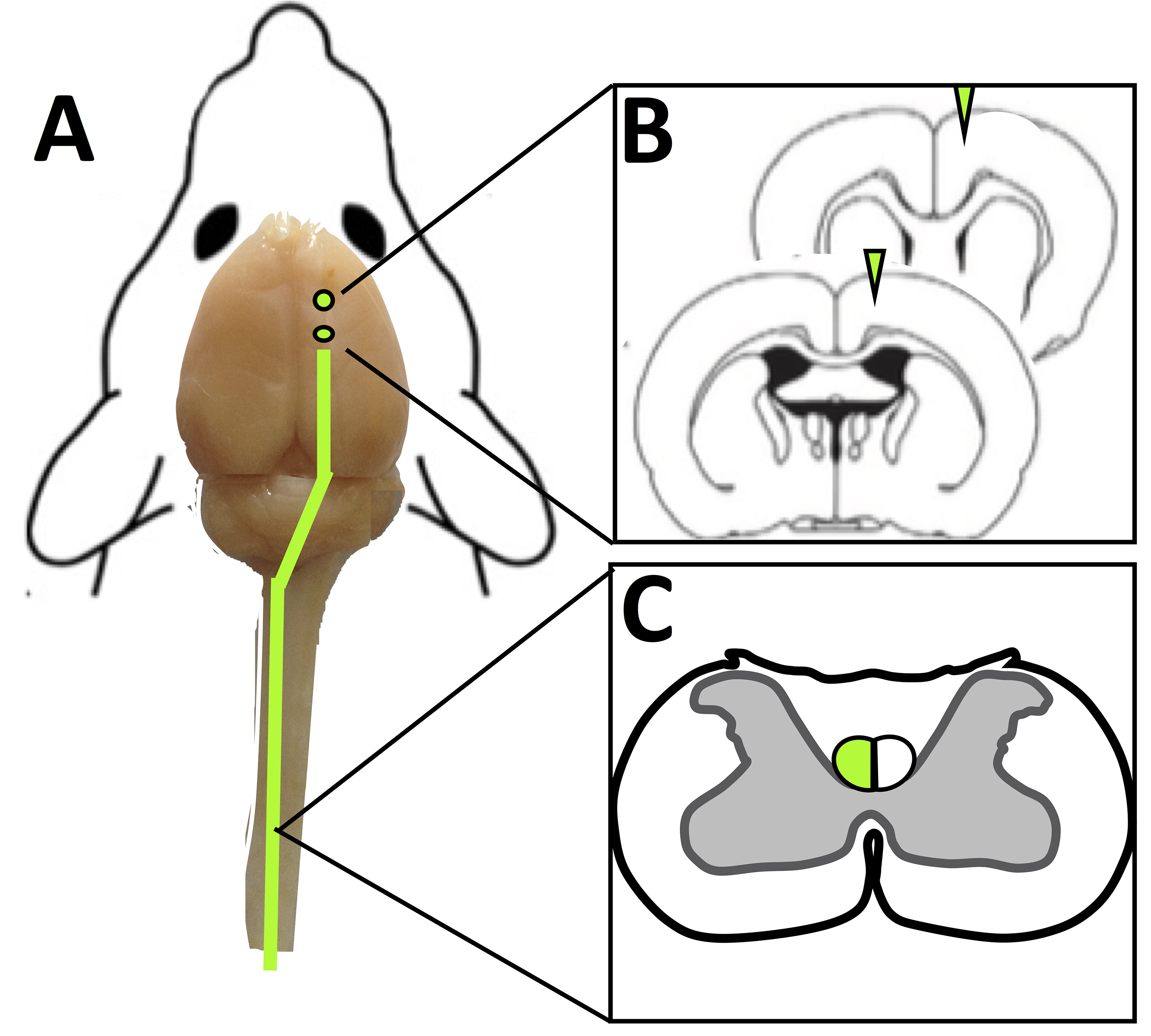
Figure 1. Use of anterograde cortical tracers to label the corticospinal tract in rats. A. Tracer injection sites in the rat cortex (shown as green circle) and anterograde tracer labeled descending fibers (green line). B. Coronal sections of the brain at tracer injection sites. C. Transverse slices of the spinal cord (C4) with tracer labeled fibers of the CST shown in green.
Materials and Reagents
- Surgical
- Sterile scalpel blade # 10 (Fine Scientific Tools, catalog number: 10100-00 )
- 10 µl Hamilton syringe (Hamilton, catalog number: 80366 )
- Glass microtubule–Glass filament with 1.0 mm external diameter (Sutter Instrument, catalog number: BF150-86-10 )
- 16-gauge needle (STEMCELL Technologies, catalog number: 28110 )
- 1 ml syringes x 5 (BD, catalog number: 309659 )
- Silk suture 5-0, P-3 reverse cutting (Ethicon, PERMAHAND®, catalog number: 640G )
- Sprague Dawley Rat, male, ~500 g,15-20 weeks of age (Charles River)
- Isoflurane USP 99% (Fresenius Kabi, catalog number: CP0406V2 )
- Compressed oxygen gas high purity 99.995% (Praxair)
- Compressed nitrous oxide gas 99% purity (Praxair)
- Sterile saline, 0.9% NaCl (Baxter, catalog number: 2B1324X )
- 0.25% Bupivacaine (Sigma-Aldrich, catalog number: B5274 )
- Buprenorphine (Schering- Plough, 0.2 mg injectable)
- Sterile scalpel blade # 10 (Fine Scientific Tools, catalog number: 10100-00 )
- Tracer
- Dextran, Alexa FluorTM 488, 10,000 MW, Anionic, Fixable (Thermo Fisher Scientific, catalog number: D22910 )
- Dextran amine, Texas Red®, 10,000 MW (Vector Laboratories, catalog number: SP-1140 )
- Dextran, Alexa FluorTM 488, 10,000 MW, Anionic, Fixable (Thermo Fisher Scientific, catalog number: D22910 )
- Perfusion
- Euthasol® (VIRBAC, catalog number: 710101 )
- Celline isotonic saline solution (Fisher Scientific, catalog number: 351142-10)
Manufacturer: SCP SCIENTIFIC, catalog number: CS20310D . - Heparin (Sigma-Aldrich, catalog number: H3393 )
- Formalin 1:10 dilution, buffered (Fisher Scientific, catalog number: SF100-20 )
- 4% formalin solution (see Recipes)
- Isotonic saline heparin (5,000 IU/L) solution (see Recipes)
- Euthasol® (VIRBAC, catalog number: 710101 )
- Tissue cryosectioning
- Superfrost® Plus Gold slides (Fisher Scientific, catalog number: 22-035813 )
- Isopentane (2-methylbutane) C5H12 (Sigma-Aldrich, catalog number: 277258 )
- D-Sucrose (Fisher Scientific, catalog number: 10638403 )
- Tissue Tek® Optimal cutting temperature (OCT) compound (Sakura, catalog number: 4583 )
- VECTASHIELD (Vector Laboratories, catalog number: H-1000 )
- 30% Sucrose solution (see Recipes)
- Superfrost® Plus Gold slides (Fisher Scientific, catalog number: 22-035813 )
- Spinal cord analysis
- Rat atlas (Paxinos, George, and Charles Watson. The rat brain in stereotaxic coordinates: hard cover edition. Access Online via Elsevier, 2006. A tool by Matt Gaidica.)
Equipment
- Heating pad (Stoelting, catalog number: 50300 )
- Betadine® surgical scrub–(Fisher Scientific, catalog number: 19-027132 )
- Sterile Crile Hemostats (Fine Science Tools, catalog number: 13004-14 )
- Sterile blunt ended scissors (Fine Science Tools, catalog number: 14001-18 )
- Sterile rongeur (Fine Science Tools, catalog number: 16000-14 )
- Carbide burs drill bit (SS WHITE, catalog number: 14002-5 )
- Perfusion tray (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 11469 )
- Sterile scoop (Fine Science Tools, catalog number: 10090-13 )
- Weigh scale (Kent Scientific, catalog number: SCL-1015 )
- Animal Anesthetic System (Harvard Apparatus, catalog number: 72-6467 )
- Stereotaxic frame (KOPF INSTRUMENTS, model: Model 963 with standard accessories)
- Micropipette puller (Sutter Instrument, catalog number: P-30 )
- Surgical drill–TCM Endo III (Nouvag, catalog number: 31817 )
- Digital Peri-Star Pro-Peristaltic Pump (World precision instruments, catalog number: PERIPRO-4HS )
- Cryostat (Leica Biosystems, catalog number: CM3050 S research cryostat)
- Leica HCX PL APO L 10x 1.0NA water emersion objective lens
- Leica SP5 Confocal microscope (Leica Microsystems, model: Leica SP5 )
- Inverted microscope (Leica Microsystems, catalog number: Leica DMI6000B )
- Animal surgery stereo microscope (Leica, M-series)
Software
- Leica LAS AF Software
- ImageJ
- Prism by Graph Pad vs9
Procedure
- Anesthetic and surgical preparation
- Weigh animal and record weight.
- Place a rat in an anesthetic induction chamber.
- Adjust the oxygen flow rate to 1.5 L/min and adjust isoflurane vaporizer to 3%, gradually increase isoflurane to 5% over 2 min.
- Once the animal losses righting reflex, stop gas flow to the chamber.
- Rapidly remove the rat from the induction chamber and place in anesthetic mask or nosecone.
- Immediately redirect flow to an anesthetic nosecone, reduce oxygen flow rate to 0.3-0.4 L/min, then set nitrogen flowmeter to 0.6-.08 L/min and reduce the isoflurane to 1.5 to 2.0%.
Note: We use 1.5-2.0% isoflurane (in 70% nitrous oxide and 30% oxygen) at a flow rate of 1 L/min to maintain anesthetic plane in 500 g rats. Anesthetic and inhaled gas choices are important to consider when inducing stroke as oxygenation and vasoconstriction can alter the size of induced stroke. - Ensure the animal is in surgical plane of anesthetic as evident by a loss of reflexes, muscles relaxation and deep rhythmic breathing in accordance with IACUC protocols.
- Ensure body temperature is maintained at 37 °C with a rectal temperature probe and a heating pad.
- Apply ophthalmic ointment to both eyes to prevent damage to the cornea, and reapply every 15 min or as needed.
- Remove hair from the surgical field on the rat scalp from the ears to the eyes by cutting hair at the root right next to the skin with a pair of sterile surgical scissors.
- Prepare skin with Betadine® surgical scrub.
- Weigh animal and record weight.
- Creation of burr holes for injection of tracer
- Place the rat in a stereotactic frame, ensuring the skull is level and the ear bars are tight.
- Induce an incision line block with 0.25% Bupivacaine (do not exceed 8 mg/kg).
- Focus the surgical area under an animal surgery stereo microscope at 5x.
- Make a 1 cm incision over the scalp midline (Figure 2A), superior to the ear and inferior to the eyes.
- Expose the skull by retracting each side of the incision away from the midline to expose bregma (Figures 2B and 2C).
Note: If you have difficulty locating bregma, apply slight pressure to one of the skull plates and the sutures will become obvious as they shift under mild pressure. - Mark the stereotaxic coordinates of the forelimb motor cortex where the burr holes will be formed at (1.5, 0) and (1.5, 2) on the skull with a marker (Figures 2D and 2E).
Note: Anterograde tracers should be injected into the motor cortex to ensure labeling of the CST. - If this protocol is being used with cortical injury models such as photothrombotic stroke of the somatosensory cortex as described by Wiersma et al., 2017, place the burr holes in the corners of the existing thin window closest to the midline (1, -1) and (1, -3) to assess the spared fibers in the cortex, as these coordinates give access to spared fibers at the periphery of the stroke (Figures 2F and 2G).
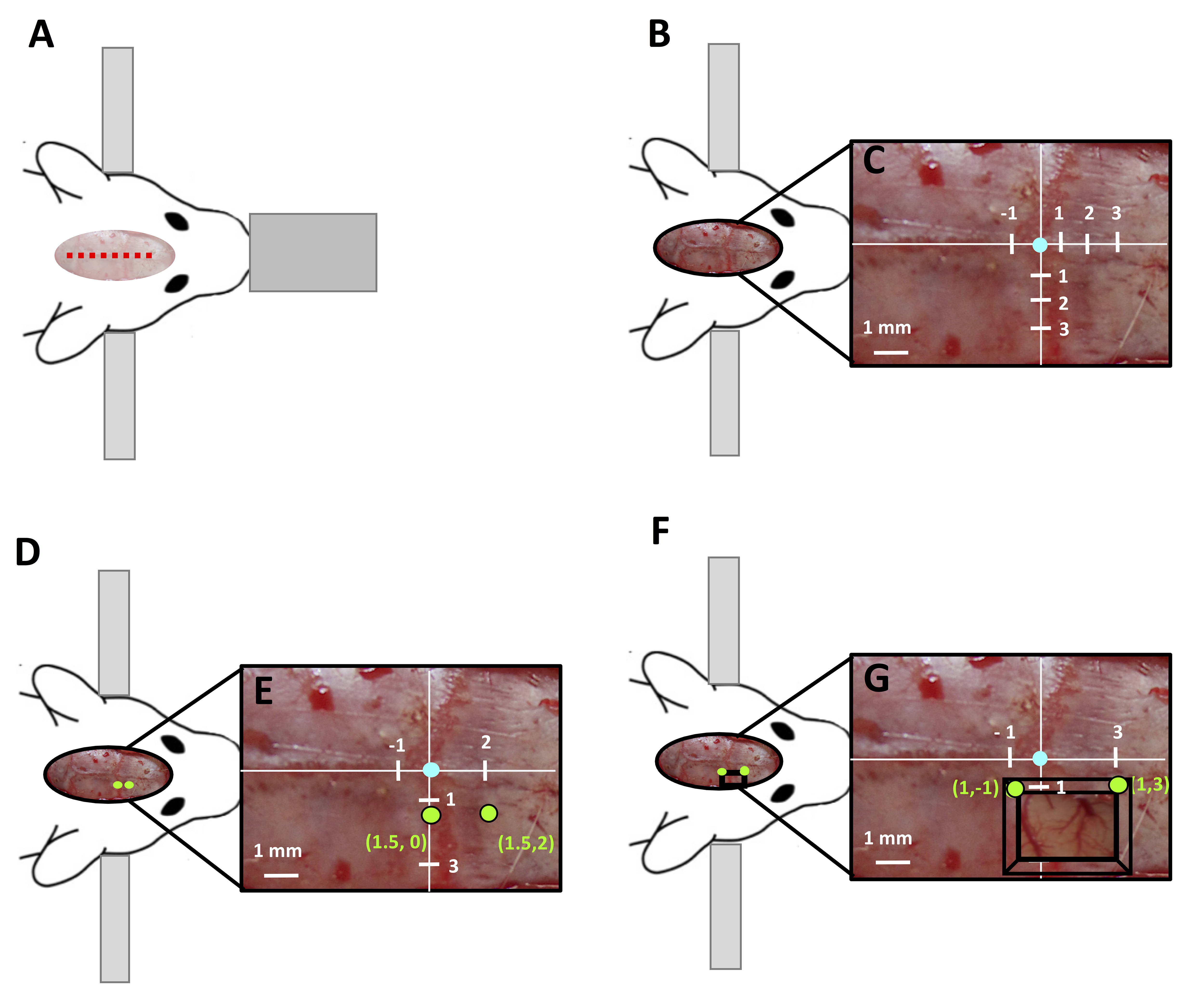
Figure 2. Creation of burr hole guided by stereotaxic coordinates. A. Surgical set up of rat in stereotaxic frame with the location of midline incision marked by red dashed line. B. Exposed skull of rat and C. stereotaxic coordinates of the skull, with bregma labeled in blue, at 5x magnification. D, E. Location of burr holes (green circle) for injection over the motor cortex with E. stereotaxic coordinates labeled in green. F, G. Location at 5x magnification. F. location of burr holes (green circle) modified for cortical injury models with a thin window (black box) such as photothrombotic stroke to label spared fibers, G. stereotaxic coordinates labeled in green) at 5x magnification. - Using a surgical drill fitted with a rounded carbide drill bit designed for cutting with a width of 1 mm, create a burr hole at each of the marked locations.
- Drill through the first 0.25 mm of the skull.
- Allow the skull to rest for 1 min–drilling creates heat which can lead to damage of the cortical surface, therefore, it is important to slowly drill the burr hole leaving time for cooling. A drop of saline can be added at this step to aid in cooling, prevent overheating of the cortical surface and to clear the viewing field.
- Repeat Steps B9 and B10 until the cortical surface becomes visible through the thinned skull such that the bottom of the burr hole will appear pink rather than white, and on the addition of a saline drop the vasculature of the underlying cortex can be seen, at this point the skull is approximately 0.25 mm thick.
Note: Expect a total skull thickness of approximately 2 mm in adult male rats. - Once the skull is thinned to 0.25 mm, it is important to drill for no more than 2 sec at a time to avoid heating of the cortical surface.
- Once the cortical surface is exposed immediately, stop drilling to avoid disruption of the dura.
- Place the rat in a stereotactic frame, ensuring the skull is level and the ear bars are tight.
- Injection of tracer
- For injection of tracer, a 10 µl Hamilton syringe fitted with a glass microtubule tip pulled to a 1 μm diameter should be used to ensure minimal damage of the cortex (Figure 3).
Note: To create a pulled tip for injection, use a 10 cm long thin-walled glass filamented microtubule with a 1.0 mm external diameter and a 0.75 internal diameter. Placed microtubule in pipette puller with a heat ramp of +10, a pull of 60-90, a velocity of 80, a delay of 90 and a pressure of 200. These settings produce constant tips with a 1 μm diameter. - Fill the syringe attached to the glass tip with 1 μl of Dextran, Alexa Fluor® 488, 10,000 MW (1 mg/μl).
Note: See information about tracer selection in the Notes section. - Mount the tracer-filled syringe on the stereotaxic arm.
- Line up the glass tip with the created burr hole.
- Lower the glass tip to the cortical surface.
- The desired injection depth of tracer is 1.4 mm below the cortical surface, therefore the Hamilton syringe is lowered via the stereotaxic arm to a depth of 1.5 mm.
Note: This should be done in one smooth motion; this is usually 1.5 full rotations of the dial on most stereotaxic arms where 1 rotation = 1 mm advancement of the stereotaxic arm. - Retract/reverse the syringe 0.1 mm.
Note: Turn the dial on the stereotaxic arm in the opposite direction for 1/10 of a rotation. This will create space for the injected tracer in the cortex. - The tracer is now ready to be injected.
- Push the plunger stem on the syringe to inject 0.2 μl.
- Wait 1 min–watching for backflow of tracer into the glass tip. If no back flow occurs, repeat the 0.2 μl injection. In the case of backflow: use the stereotaxic arm to retract/reverse the syringe 0.1 mm and wait an additional 1 min, the backflow should clear out of the glass tip back into the cortex.
- Repeat Steps C9-C10 until the entire 1 μl volume of tracer has been injected.
- The syringe can now be removed from the brain in small 0.2 mm steps.
- Reverse the syringe via the stereotaxic arm 0.2 mm.
- Wait 30 sec.
- Repeat Steps C13 and C14 until the syringe is out of the cortex.
- Repeat Steps C1-C15 in the second burr hole.
- Suture the rat scalp and give analgesic care in accordance with IACUC protocols (0.5 mg/kg subcutaneous buprenorphine).
- Give a subcutaneous injection of 3 ml saline to prevent dehydration.
- Immediately after surgery, observe the animal for signs of paralysis and weakness in the forelimb as this is an indication that the burr hole or injection has induced unwanted damage to the cortex.
- For injection of tracer, a 10 µl Hamilton syringe fitted with a glass microtubule tip pulled to a 1 μm diameter should be used to ensure minimal damage of the cortex (Figure 3).
- Spread of tracer for cortex to spinal cord
- Allow the animal to recover for 14 days after tracer injection. This will ensure sufficient time for the tracer to travel from the cortex to the spinal cord.
Note: We found optimal labeling of the CST in the spinal cord 14 days after cortical injections.
- Allow the animal to recover for 14 days after tracer injection. This will ensure sufficient time for the tracer to travel from the cortex to the spinal cord.
- Transcardial perfusion of rat
- Prepare perfusion solutions (4% formalin and Saline with heparin) and allow time to reach correct temperature for use (Recipe 1).
- Intraperitoneally inject Euthasol® (150 mg/kg) in accordance with the American Veterinary Medical Association Guidelines for Euthanasia.
- Wait for surgical plane to be reached then secure animal on its back to a perfusion tray set at 85°.
- Locate the xiphoid process following the xiphoid plane make a transverse incision through the integument and abdominal wall following just beneath the rib cage.
- Make an incision through the diaphragm using blunt-ended scissors and continue the incision along the entire rib cage exposing the pleural cavity.
- Make sagittal incisions through both sides of the rib cage, ensuring not to damage the heart or lungs.
- Pin the rib cage to reveal the coronal chest and release the heart of any attachments to the chest cavity.
- Using a hemostat grip the apex of the heart and insert a blunt-ended 16 gauge needle through the left apex into the left ventricle.
- Make a small incision in the right atria to release fluid pressure and start primed peristaltic perfusion pump at 70-100 ml/min. Flush 250 ml of 37 °C isotonic saline Heparin (5,000 IU/L) solution (Recipe 2).
Note: The animal should become pale, the liver should be white and only clear fluids should be draining from the right atrial in successful perfusions. - Switch perfusion fluid to 4 °C 4% formalin solution, again perfuse 250 ml of the solution at a rate of 70-100 ml/min.
Note: Fixative tremors should begin to occur through the rat within seconds of the 4% formalin solution being infused and the rat should be very stiff by the end of the perfusion.
- Prepare perfusion solutions (4% formalin and Saline with heparin) and allow time to reach correct temperature for use (Recipe 1).
- Collection of rat spinal cord
- Create a midline incision from the tail to the top of the skull between the eyes.
- Remove the skin and muscle covering the skull and spinal cord.
- Starting at the lower lumbar spinal cord, use a rongeur to cut the superior vertebral notch of the vertebra releasing the vertebral body from the laminae, transverse processes and the spinous process. The spinal cord will adhere to the vertebral body of the now open spinal canal.
- Continue to open the spinal canal of each vertebra from the lumbar to cervical spinal cord.
- The rat skull can now be removed by sliding a small blunt scissor under the skull along the midline. The scissors can be used to peel the skull plates to each side exposing the brain.
- Once the brain and spinal cord are exposed, begin removing the spinal cord from the open spinal cavity.
- Starting at the lumbar spine, lift the spinal cord out of the open spinal canal, making sure to cut the dorsal roots as they may prevent the spinal cord from being removed.
- Continue releasing the spinal cord, lifting it up as you go.
Note: The spinal cord is very delicate at this point, do not squeeze any section or pull with too much force as it may break. Do not let the spinal cord touch any surrounding tissue as it is very sticky and can easily adhere to any surface it touches. - When the spinal cord is fully released, remove the attached brain.
Note: The brain does not need to be removed with the spinal cord, however, when attached it weights down the spinal cord during post fixing preventing twisting. - Insert a rongeur into the void where the cervical vertebra has been detached sliding it along the occipital skull.
- Cut through the occipital skull and pull the bone away from the hindbrain, repeat on the other side.
- Insert a blunt scissor under the skull along the midline, cutting the skull at the midline.
- Peel each half of the skull away from the brain.
- Clip any sharp edges remaining as the brain is now ready to be removed from the open skull cavity.
- Insert a small scoop between the frontal lobe and olfactory bulbs releasing the brain from the bulbs.
- Run the scoop under the brain until all connecting tissues and blood vessels are cleared and the brain easily slides out of the open skull.
- Create a midline incision from the tail to the top of the skull between the eyes.
- Tissue fixing, freezing and cryosectioning
- Immediately following removal, place the brain and spinal cord in 50 ml of 4% formalin, leave overnight at 4 °C.
- After overnight fixation, replace 4% formalin with 50 ml of 4 °C 30% sucrose solution (Recipe 3) and leave tissue for 3-4 days at 4 °C until the brain is no longer buoyant in solution.
- Remove tissue from solution (tissue will be stiff and white in color).
- The brain and spinal cord can now be separated.
- Cut a piece of filter paper piece of filter paper that is 2 cm longer and 4 cm wider than the spinal cord.
- Lay the spinal cord flat on the OCT covered filter paper; OCT should help to ensure the spinal cord does not twist while freezing.
- If the spinal cord does not lay flat on the OCT layer, the OCT can be chilled to decrease the viscosity and provide a more ‘sticky’ layer which will force the spinal cord to lay flat on the filter paper.
- Submerge the spinal cord lying flat on filter paper in 2-methylbutane solution chilled to -50 °C.
- Leave the cord submerged for 1 min to ensure it is frozen throughout.
Note: If the spinal will be stored for long periods of time (more than 2 weeks) the tissue should be coated in an additional layer of OCT freezing media. - The tissue is now ready for long-term storage at -20 °C or immediate cryosectioning.
Note: If planning to section the tissue, we do not recommend storage at -80 °C, as the tissue must be warmed to -20 °C for sectioning. We experienced fracturing of tissues and the appearance of freeze-thaw line when we warmed our tissue to optimal sectioning temperature after storage at -80 °C). - A cryostat should be set to a temperature between -19 and -22 °C, with a cutting thickness of 35 μm for transverse sections of the spinal cord.
- Cut the spinal cord into 10 mm sections which can then be mounted with OCT for sectioning. Starting cutting the 10 mm segments from the superior cervical spinal cord where the brain stem has been removed from the spinal cord and continue through to the inferior end of the lumbar spinal cord.
Note: Spinal cord segments longer than 10 mm tend to vibrate while sectioning leading to cracking of the tissue. - The spinal cord can now be sectioned in 30 μm slices and mounted onto glass microscope slide.
Note: We recommend Superfrost® Plus Gold slides as spinal cords have a small surface area to adhere to the slide with, which can result in tissue slices lifting and floating off during processing. - Once slices are mounted then should warmed to 4 °C.
- Cover slide with 2 drops of VECTASHIELD mounting media to preserve fluorescents of the tracer.
- Store slides at 4 °C out of direct light as UV light exposure can bleach the fluorescent tracer.
Note: Fluorescent tracers will fade over time, tissue should be analyzed within 28 days of sectioning and mounting to avoid degradation of the tracer.
- Immediately following removal, place the brain and spinal cord in 50 ml of 4% formalin, leave overnight at 4 °C.
- Fluorescent microscopy
- Tissue was analyzed on a Leica SP5 Confocal microscope using a Leica HCX PL APO L 20 x 1.0NA water emersion objective lens and Leica LAS AF Software.
- Visualize AlexaFluor 488-tagged dextran tracers using spectral settings for AlexaFluor 488 or FITC emission spectrum on a confocal microscope.
- Collect 30 μm z-stacks composed of 10 x 0.3 μm steps taken at 600 Hz over a 736 x 736 μm section of tissue.
- Set up the 736 x 736 tile scan to span across the grey matter of both the ventral and dorsal horns as well as the CST to on the spinal cord slice to be analyzed.
- Collect images from 20 spinal cord sections for each spinal segment.
Note: Analysis is shown for segments C4 and C5 as this is where the forelimb motor innervation branches off in the spinal cord (Figures 3, 4), other areas of the spinal cord can be analyzed and tracer can be visualized throughout the spinal cord including the lumbar segments. The rat atlas can be used to aid identifying which spinal segment is being assessed.
- Tissue was analyzed on a Leica SP5 Confocal microscope using a Leica HCX PL APO L 20 x 1.0NA water emersion objective lens and Leica LAS AF Software.
- Neuroanatomical tracer quantification
- Open the collected images in ImageJ (Figure 3A).
- Have a blind observer count and record the number of labeled fibers in the CST of each section.
Note: A fiber is considered positive for labeling if it is visible in green using spectral settings for AlexaFluor 488 or FITC emission spectrum on a confocal microscope. See Figures 3B and 3C for location of CST and examples of positively labeled fibers. - Have a blind observer count and record the number of labeled projections into the grey matter on the traced side of the spinal cord (Figures 3D and 3E).
- Have the observer measure the length of each axon segment greater than 1 μm counted in Step I3.
- Have a blind observer count and record the number of projections into the grey matter on the untraced side of the spinal cord (Figures 3F and 3G).
- Have the observer measure the length of each axon segment greater than 1 μm counted in Step I5.
- Repeated on 20 spinal cord sections per segment analyzed.
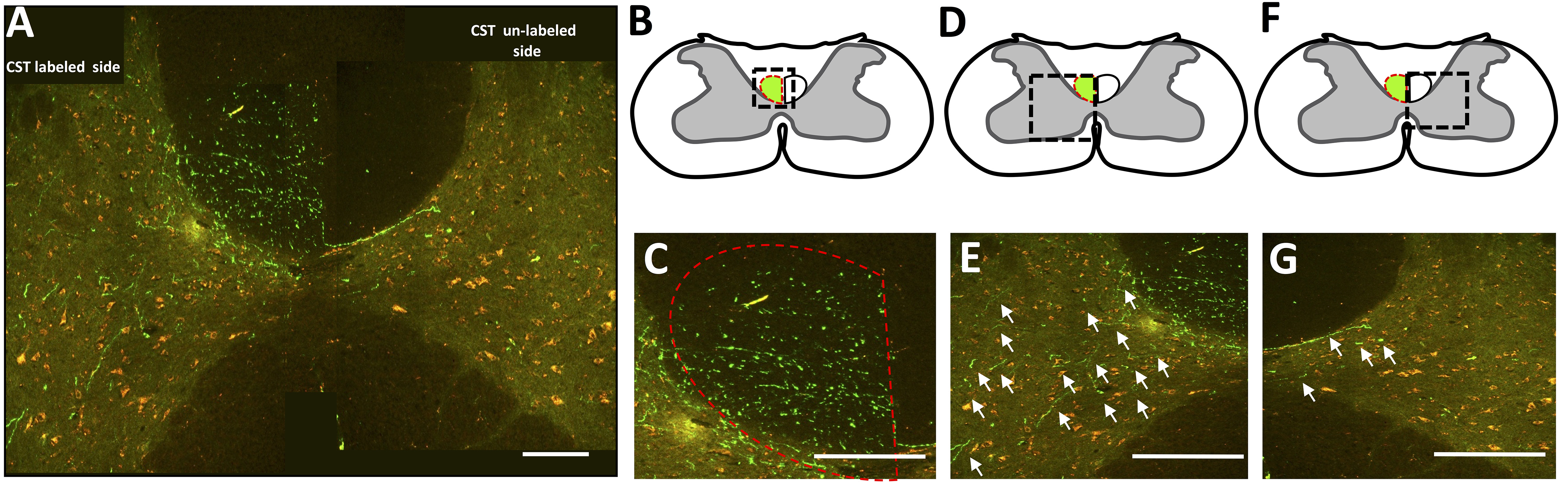
Figure 3. Analysis of labeled CST fibers in the spinal cord. A. Tile scan image of tracer labeled CST and surrounding grey matter in the cervical spinal cord (C4). B. The CST tract of a transverse spinal slice is outlined in red, the black dashed box is enlarged in C. showing many labeled CST fibers in green. D. The grey matter adjacent to the labeled CST is outlined in black dashed lines and shown in E. where fibers in the grey matter on the labeled side of the spinal cord that should be counted are pointed out with white arrow. F. The grey matter contralateral to the labeled CST is outlined in black dashed box which is shown in G. where some fibers in grey matter on the unlabeled side of the spinal cord that should be counted are pointed out with white arrow. Scale bars = 2 mm, adapted from Wiersma et al., 2017.
- Open the collected images in ImageJ (Figure 3A).
- Neuroanatomical tracer fiber length analysis
- Add together the number of labeled fibers counted in the CST in each of the 20 analyzed slices.
- Add the length of all the axon collaterals measured in the 20 slices on the tracer-labeled side of the spinal cord (Step I4).
- Divide the sum of the axon collateral length of the 20 slices (Step I2) by the total number of axons counted in the CST of the 20 slices to determine the average length of traced fibers on the labeled side of the spinal cord.
Note: This reduces variability due to inter-animal differences in tracer efficacy or uptake.
- Add together the number of labeled fibers counted in the CST in each of the 20 analyzed slices.
- Stereological quantification of fibers sprouting
- Fit a 50 x 40 grid over the grey matter (Figure 4A).
- Position dorsal to ventral lines from the tip of lamina to the border of ventral horn grey matter.
- Position medial to lateral lines from the midline the lateral grey matter border (Figure 4A).
- The grid is custom fitted over the grey matter to ensure that variation in tissue shrinkage, orientation and deformation would not affect the total fiber counts (Figure 4B).
- To create heat plots, count the number of fiber in each grid section and normalize to the average number of fibers counted in the grid sections of the labeled CST.
- Assign a color to each section of the grid based on the average number of fibers counted in that section over the 20 sections (Figure 4C).
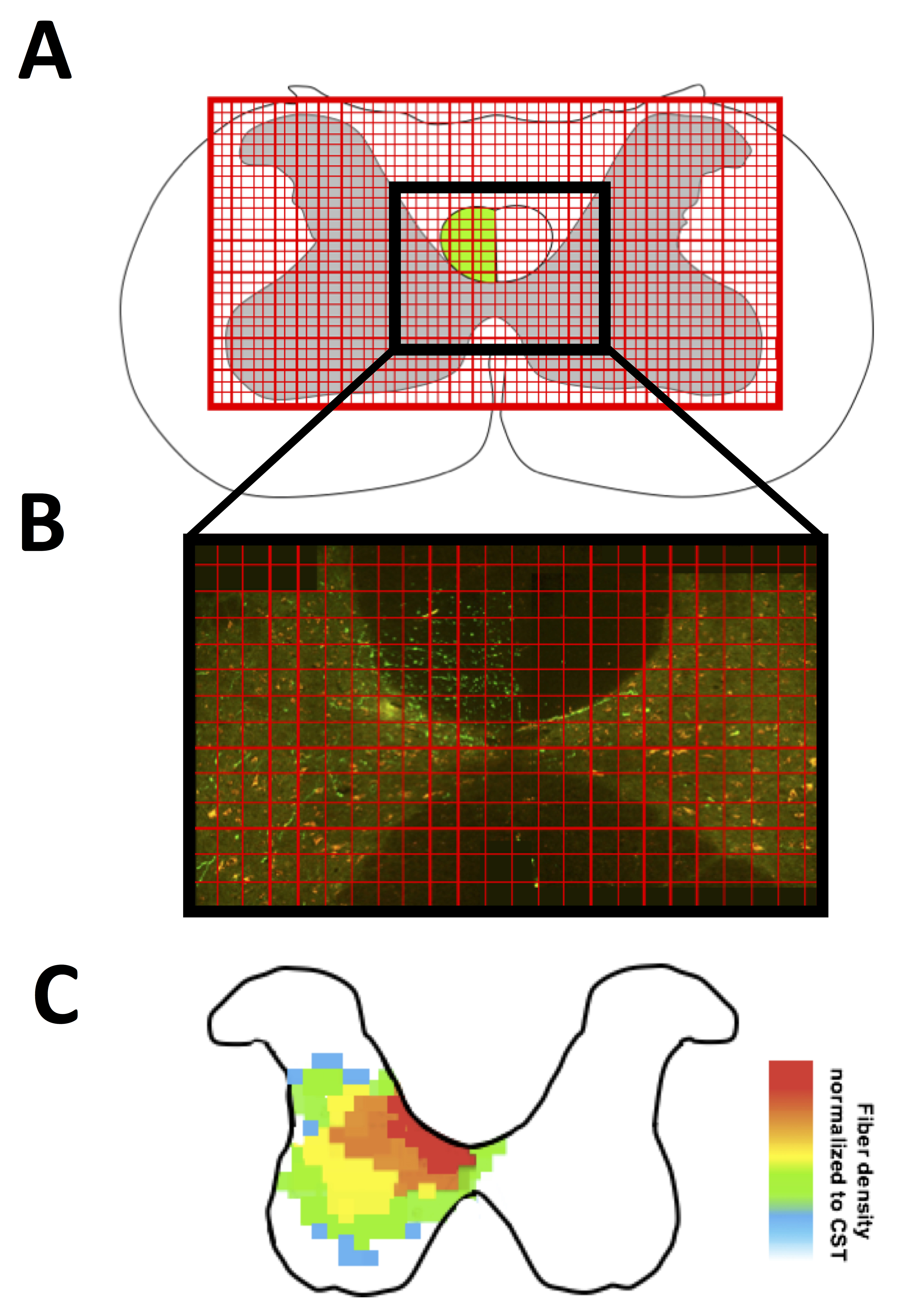
Figure 4. Stereological quantification of fibers sprouting. A. Example of grid applied over the grey matter of a transverse section; B. Application of grid over the CST on image; C. Example heat map. Adapted from Wiersma et al., 2017.
- Fit a 50 x 40 grid over the grey matter (Figure 4A).
Data analysis
To determine if the number of labeled fibers in the CST is significantly different between two groups, use Student’s unpaired two-tailed t-test. To analyze statistical differences between groups in fiber count and length, use two-way ANOVA testing followed by post hoc Holm–Sidak’s testing.
Notes
- In the case of surgical complications consult the American Veterinary Medical Association Guidelines for Euthanasia (2013) for a comprehensive guideline on evaluating decisions regarding euthanasia and the morality of such decisions.
- Tracer selection is very important, dextran tracers of 10,000 MW will allow axons and terminals of the central nervous system to be resolved, however, if labeling of the peripheral nervous system or cell bodies is required, select a tracer of 3,000 MW.
- Tracers can be injected into the sensory-motor cortex of both cortices to assess the CST descending from both sides of the cortex. We have found that using Dextran, Alexa FluorTM 488, 10,000 MW, Anionic, Fixable (Thermo Fischer) on one side of the cortex and Dextran amine, Texas Red®, 10,000 MW (Vector Laboratories) on the other side of the cortex yield good results.
- The methods of anterograde tracing presented here can easily be combined with the photothrombotic stroke model, where the burr holes for tracer injection are made in the corners of the thin window designed for induction of photothrombosis of the somatosensory forelimb cortex. Tracers injected in the ipsilesional to the infract allow assessment of spared fibers in the contralesional spinal cord.
- This protocol can also be used with adeno-associated viral vectors (AAVs) expressing green fluorescent protein (GFP) or mCherry (a red fluorescent protein) to label CST axons originating in ipsilesional or contralesional motor cortex. Through the burr hole 1 µl AAV solution (~108 transducing units per microliter of AAV solution) expressing either GFP (AAV1. CB7.CI.eGFP.WPRE.rBG,UPenn Vector Core) or mCherry (AAV1.CB7.CI.mCherry.WPRE.rBG) is injected into cortical layers V and VI at two locations immediately medial to the stroke (to label any spared CST fibers in the primary motor cortex) and homotypical motor cortex contralateral to the lesion (to label uninjured fibers). These tracers were selected over conventional dextran-based tracers because they offer improved signal to noise labeling in anterograde fibers with more robust precisely defined injection sites.
- ANOVA testing revealed no significant within-group (intragroup) variation (n = 10) in the number of labeled CST fibers, the number of projecting fibers or the length of projecting fibers. All animals had CST labeled fibers counts within one standard deviation of the mean, we found very consistent labeling of fibers in each experimental group. In intact animals (n = 10) had an average of 111 tracer labeled CST fibers at C4 with a standard deviation of 38 and a standard mean error of 13.45.
Recipes
- 4% (1:25) formalin solution (500 ml)
- Measure 300 ml isotonic saline
- Add 200 ml of 1:10 formalin solution
- Invert to mix
- Chill to 4 °C
- Measure 300 ml isotonic saline
- Isotonic saline heparin (5,000 IU/L) Solution (500 ml)
- Measure 500 ml isotonic saline
- Warm saline to 37 °C
- Add 2,500 IU heparin
Note: Heparin will denature overtime at 37 °C, do not add until right before use. - Invert to mix heparin into saline
- Measure 500 ml isotonic saline
- 30% sucrose solution (50 ml)
- Measure 45 ml of isotonic saline
- Add 15 g of D-sucrose
- Mix vigorously
- Chill to 4 °C
- Measure 45 ml of isotonic saline
Acknowledgments
This work was supported by Alberta Innovates Health Solutions, Heart and Stroke Foundation of Canada, Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada. Grants from the Canada Foundation for Innovation and Province of Alberta Small Equipment Grants were used to acquire confocal microscopy equipment. This protocol and figures are adapted from previous work: Wiersma, A. M., Fouad, K. and Winship, I. R. (2017). Enhancing spinal plasticity amplifies the benefits of rehabilitative training and improves recovery from stroke. J Neurosci 37(45): 10983-10997. The authors have no conflicts of interest to declare.
References
- American Veterinary Medical Association Guidelines for Euthanasia (2013) (AVMA).
- Hill, C. E., Beattie, M. S. and Bresnahan, J. C. (2001). Degeneration and sprouting of identified descending supraspinal axons after contusive spinal cord injury in the rat. Exp Neurol 171(1): 153-169.
- Starkey, M. L., Bartus, K., Barritt, A. W. and Bradbury, E. J. (2012). Chondroitinase ABC promotes compensatory sprouting of the intact corticospinal tract and recovery of forelimb function following unilateral pyramidotomy in adult mice. Eur J Neurosci 36(12): 3665-3678.
- Thallmair, M., Metz, G. A., Z'Graggen, W. J., Raineteau, O., Kartje, G. L. and Schwab, M. E. (1998). Neurite growth inhibitors restrict plasticity and functional recovery following corticospinal tract lesions. Nat Neurosci 1(2): 124-131.
- Ueno, M., Hayano, Y., Nakagawa, H. and Yamashita, T. (2012). Intraspinal rewiring of the corticospinal tract requires target-derived brain-derived neurotrophic factor and compensates lost function after brain injury. Brain 135(Pt 4): 1253-1267.
- Vavrek, R., Girgis, J., Tetzlaff, W., Hiebert, G. W. and Fouad, K. (2006). BDNF promotes connections of corticospinal neurons onto spared descending interneurons in spinal cord injured rats. Brain 129(Pt 6): 1534-1545.
- Wahl, A. S., Omlor, W., Rubio, J. C., Chen, J. L., Zheng, H., Schroter, A., Gullo, M., Weinmann, O., Kobayashi, K., Helmchen, F., Ommer, B. and Schwab, M. E. (2014). Neuronal repair. Asynchronous therapy restores motor control by rewiring of the rat corticospinal tract after stroke. Science 344(6189): 1250-1255.
- Wiersma, A. M., Fouad, K. and Winship, I. R. (2017). Enhancing spinal plasticity amplifies the benefits of rehabilitative training and improves recovery from stroke. J Neurosci 37(45): 10983-10997.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Wiersma, A. M. and Winship, I. R. (2018). In vivo Use of Dextran-based Anterograde Cortical Tracers to Assess the Integrity of the Cortical Spinal Tract. Bio-protocol 8(10): e2862. DOI: 10.21769/BioProtoc.2862.
- Wiersma, A. M., Fouad, K. and Winship, I. R. (2017). Enhancing spinal plasticity amplifies the benefits of rehabilitative training and improves recovery from stroke. J Neurosci 37(45): 10983-10997.
Category
Neuroscience > Nervous system disorders > Animal model
Neuroscience > Neuroanatomy and circuitry > Animal model
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


.jpg)






