- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enzymatic Activity Assay for Invertase in Synechocystis Cells
Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2856 Views: 7722
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3552 Views

Activation of X-Succinate Synthases for Fumarate Hydroalkylation Using an In Vitro Activation Method
Anshika Vats [...] Mary C. Andorfer
Jun 20, 2025 2511 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2442 Views
Abstract
Invertase can catalyze the hydrolysis of sucrose, and is widely distributed in cells of cyanobacteria and plants. Being responsible for the first step for sucrose metabolism, invertase plays important physiological roles and its enzymatic activity is frequently needed to be determined. All the methods for determination of the invertase activity are dependent on detection of the glucose product generated by the invertase. Here we describe an ion chromatography based protocol of our laboratory for determination of cyanobacterial intracellular invertase activity.
Keywords: CyanobacteriaBackground
Invertase and sucrose play important physiological roles in cyanobacteria (Curatti, et al., 2008; Kolman et al., 2015) and higher plants (Vargas et al., 2003; Vargas et al., 2010). Invertase (EC 3.2.1.26) can catalyze the sucrose degradation into glucose and fructose. Due to this characteristic of the invertase, any methods which could be used for the determination of glucose or fructose would be theoretically used for the invertase enzymatic activity assay. In fact, most of the invertase enzymatic activity assays are based on the detection of the generated glucose product.
Some companies have developed several kits for the invertase activity assay, for instances, ab197005 from abcam (USA), KA1629 from Novus Biologicals (USA), MAK118 from Sigma-Aldrich (USA). By using these kits, the glucose product generated from the invertase reactions would be oxidized and determined by a colorimetric (570 nm) or fluorimetric method (λem/ex = 585/530 nm). In other methods, the amount of reducing sugar liberated by invertase was measured by coupling hexokinase, phosphoglucose isomerase and glucose-6-phosphate dehydrogenase, while the resulting NADPH was further spectrophotometrically determined at 340 nm (Vargas et al., 2003). Ion chromatography could directly detect various sugars including glucose, fructose, sucrose (Du et al., 2013) and glucosylglycerol (Tan et al., 2015). Compared with the spectrophotometer-based methods for sugar determinations, ion chromatography (IC) could be more beneficial for analyzing the enzymatic mixtures containing multiple compounds, especially for the enzymatic assay of cell crude extracts. By using ion chromatography, the sucrose consumption and the glucose production could be shown at the same time in the case of the invertase enzymatic assay, which would provide complete information for the enzymatic assays.
Here, we report our recent IC-based protocol for determination of the invertase activity in cyanobacterial cells.
Materials and Reagents
- Pipette tips
- 2 ml tubes (Cypress, China)
- 10 ml conical tubes (Kangjian, China)
- 1 ml syringe (Jianshi, China)
- Syringe membrane filters, 0.22 μm (Jinteng, China)
- Synechocystis sp. PCC 6803 (Tan et al., 2011)
- Milli-Q water (Millipore, Germany)
- Glass beads (Sigma-Aldrich, catalog number: G9018-250G )
- Fructose standard (Sinopharm Chemical Reagent, catalog number: 63003034 )
- Glucose standard (Sinopharm Chemical Reagent, catalog number: 10010518 )
- Liquid nitrogen
- 200 mM NaOH (prepared with Milli-Q water)
- Sucrose (Sinopharm Chemical Reagent, catalog number: 10021418 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sinopharm Chemical Reagent, catalog number: 10013018 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sinopharm Chemical Reagent, catalog number: 20011160 )
- Critic acid monohydrate (C6H8O7·H2O) (Sinopharm Chemical Reagent, catalog number: 10007118 )
- Ferric ammonium citrate ((NH4)3FeC12H10O14/C6H8O7·xFe3·yNH3) (Sinopharm Chemical Reagent, catalog number: 30011428 )
- EDTA·2Na·2H2O (Sinopharm Chemical Reagent, catalog number: 10009717 )
- Sodium carbonate (Na2CO3) (Sinopharm Chemical Reagent, catalog number: 10019260 )
- Boric acid (H3BO3) (Sinopharm Chemical Reagent, catalog number: 10004818 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Sinopharm Chemical Reagent, catalog number: 20026118 )
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sinopharm Chemical Reagent, catalog number: 10024018 )
- Sodium molybdate dihydrate (Na2MoO4·2H2O) (Sinopharm Chemical Reagent, catalog number: 10019818 )
- Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sinopharm Chemical Reagent, catalog number: 10008218 )
- Cobalt(II) chloride hexahydrate (CoCl2·6H2O) (Sinopharm Chemical Reagent, catalog number: 10007216 )
- Sodium nitrate (NaNO3) (Sinopharm Chemical Reagent, catalog number: 10019918 )
- Potassium dihydrogen phosphate (KH2PO4) (Sinopharm Chemical Reagent, catalog number: 10017618 )
- Dipotassium hydrogen phosphate trihydrate (K2HPO4∙3H2O) (Sinopharm Chemical Reagent, catalog number: 10017518 )
- Phenylmethanesulfonyl fluoride (Sigma-Aldrich, catalog number: P7626 )
- Isopropanol (Sinopharm Chemical Reagent, catalog number: 40064360 )
- BG11 medium (see Recipes)
- 100 mM potassium phosphate buffer (pH 7.0) (see Recipes)
- 100 mM PMSF (see Recipes)
Equipment
- 50 ml flasks
- Pipettes (Eppendorf, Germany)
- Shaker (Taicang Huamei, model: THZ-701B )
- Water bath (Shanghai Yarong, model: B-260 )
- Centrifuge (Beckman Coulter, model: Microfuge® 22R )
- -20 °C freezer (Haier, model: BCD-219D )
- Vortex-Genie 2 (Scientific Industries, model: Vortex-Genie 2 )
- Thermal Cycler for PCR (Bio-Rad Laboratories, model: T-100 )
- Ion chromatography (Thermo Fisher Scientific, Thermo ScientificTM, model: DionexTM ICS-5000+ )
- DionexTM CarboPacTM PA10 analytical column (4 x 250 mm, Thermo Fisher Scientific, model: DionexTM CarboPacTM PA10 )
Software
- ChromeleonTM software (Thermo Fisher Scientific, Thermo ScientificTM, version 6.80; catalog number: CHROMELEON6)
Procedure
- Cultivation of Synechocystis sp. PCC 6803 (Tan et al., 2011)
Note: Monitor the growth of cyanobacterial cells by measuring the optical density (OD) at 730 nm with a spectrophotometer. Culture volume should be less than 30 ml in 50 ml flasks.- Grow Synechocystis cells in 20 ml liquid BG-11 media (see Recipe 1) in 50 ml flasks with an initial OD730 of 0.05.
- Cultivate the Synechocystis cultures at 30 °C with shaking (150 rpm) and constant white light illumination (50 μE/m2/sec) for ~4 days to reach the exponential growth phase.
- Grow Synechocystis cells in 20 ml liquid BG-11 media (see Recipe 1) in 50 ml flasks with an initial OD730 of 0.05.
- Preparation of cell crude extracts from Synechocystis cells
- Collect 8 ml of the exponential phase culture (OD730 ≈ 2) of Synechocystis by centrifugation at 8,000 x g for 10 min at room temperature.
- Resuspend Synechocystis cells in 0.5 ml of 100 mM potassium phosphate buffer (pH 7.0) in a 2 ml tube.
- Add 1 g of glass beads (150-212 μm; Sigma-Aldrich) into the 2 ml tube containing cell suspensions and 100 mM Phenylmethanesulfonyl fluoride (PMSF) to reach a final concentration of 1 mM. Then, vortex the tube at the highest speed for 1 min. For each 10 sec vortexing, cool the tube on ice for 5 sec.
- After centrifugation at 12,000 x g, 4 °C for 30 min, transfer the supernatant into a new tube and store on ice before use.
Note: The cell crude extracts should be used immediately.
- Collect 8 ml of the exponential phase culture (OD730 ≈ 2) of Synechocystis by centrifugation at 8,000 x g for 10 min at room temperature.
- Detection of the in vitro invertase activity
- For 50 µl in vitro invertase reactions, mix potassium phosphate buffer (pH 7.0), sucrose and crude extracts together (The final concentrations of potassium phosphate and sucrose are 100 mM and 2 g/L respectively. Whereas, the protein concentrations of crude extracts are 2.5~4.0 g/L.) Use the reaction mixture without the cell crude extract as a negative control.
- Incubate the mixtures at 30 °C in a thermal cycler (T100, Bio-Rad, USA) for 1.5 h. Then, stop the reaction by three freeze-thaw cycles. In each freeze-thaw cycle, freeze the reaction mixtures in liquid nitrogen and next thaw by incubation in a 65 °C water bath. Measure the amount of the resulting glucose and fructose as well as the rest of the sucrose substrate using ion chromatography (Figure 1).
- Spin down the reaction mixtures at 12,000 x g for 10 sec. Then, subject 25 μl of the diluted mixtures to ICS-5000+ ion-exchange chromatography system equipped with an electrochemical detector and a DionexTM CarboPacTM PA10 analytical column (4 x 250 mm, ThermoFisher, Waltham, MA, USA). Elute the column with 200 mM NaOH at a flow rate of 1.0 ml/min.
Note: The reaction mixtures should be diluted to at least 50 folds before analyzed by ion chromatography. - Prepare a series of glucose standard solutions (0.01, 0.05, 0.1, 0.5, 1, 2 mg/L), establish the standard curve by using ion chromatography (see below).
- For 50 µl in vitro invertase reactions, mix potassium phosphate buffer (pH 7.0), sucrose and crude extracts together (The final concentrations of potassium phosphate and sucrose are 100 mM and 2 g/L respectively. Whereas, the protein concentrations of crude extracts are 2.5~4.0 g/L.) Use the reaction mixture without the cell crude extract as a negative control.
- Quantification of glucose resulted from the sucrose degradation catalyzed by invertase
- Retention times of glucose and fructose using the method described above are 3.5 and 3.8 min respectively (Figure 1).
- Establish the glucose standard curve by using the peak area calculated by the Chromeleon software (Figure 2).
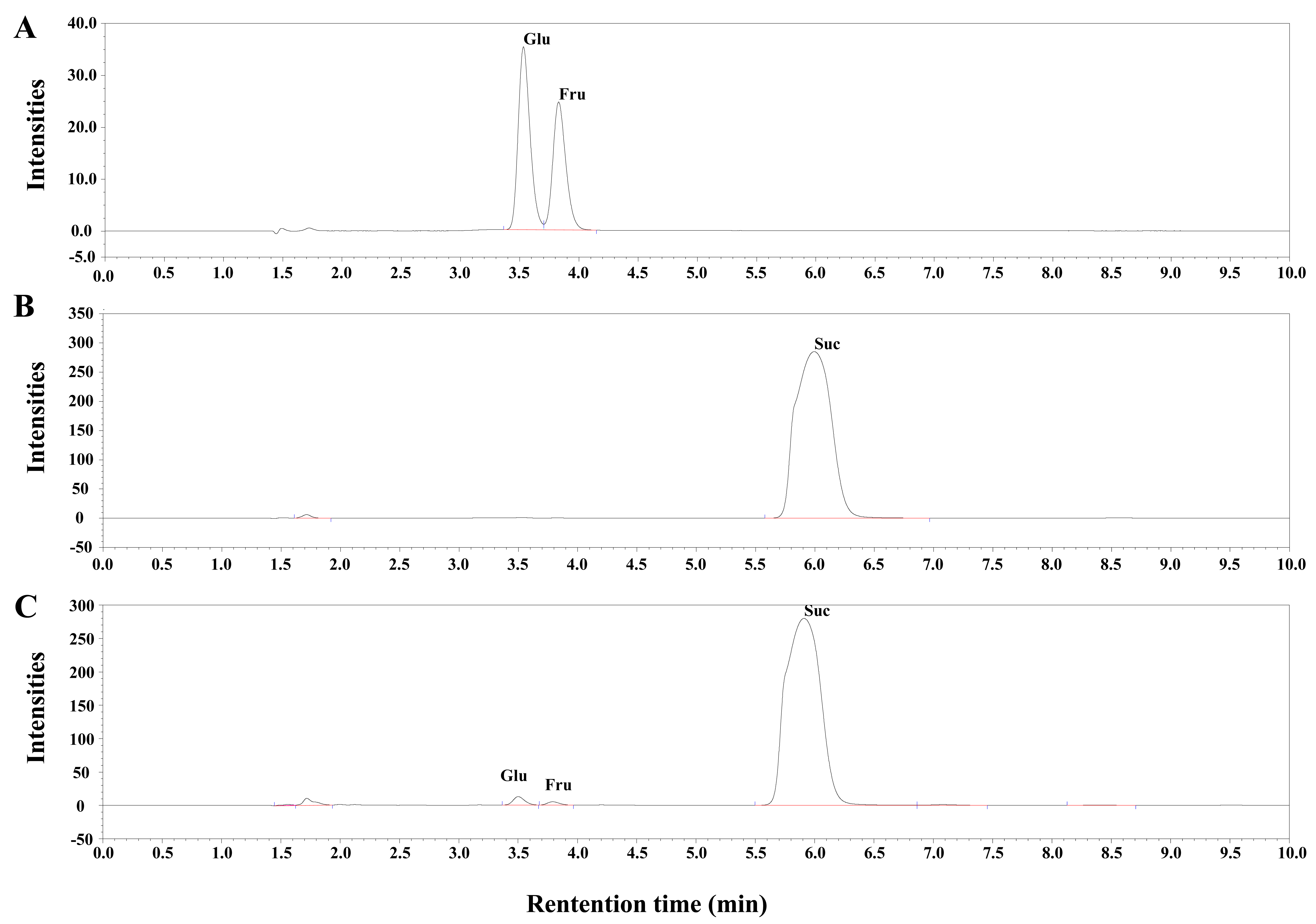
Figure 1. Ion chromatography profiles of standards and the in vitro invertase reaction mixtures. A. Chromatogram of the glucose-fructose standard mixture. 100 mg/L of the glucose and fructose standards were mixed together and analyzed by ion chromatography. B. Chromatogram of the in vitro invertase reaction mixture without the cell crude extract (negative control). C. Chromatogram of the in vitro invertase reaction mixture. Glu, Glucose; Fru, Fructose; Suc, Sucrose. - For quantification of glucose in reaction mixtures, calculate the areas of the target peak obtained by ion chromatography by the Chromeleon software, and then determine the glucose concentration using the standard curve of the glucose standards (Figure 2).
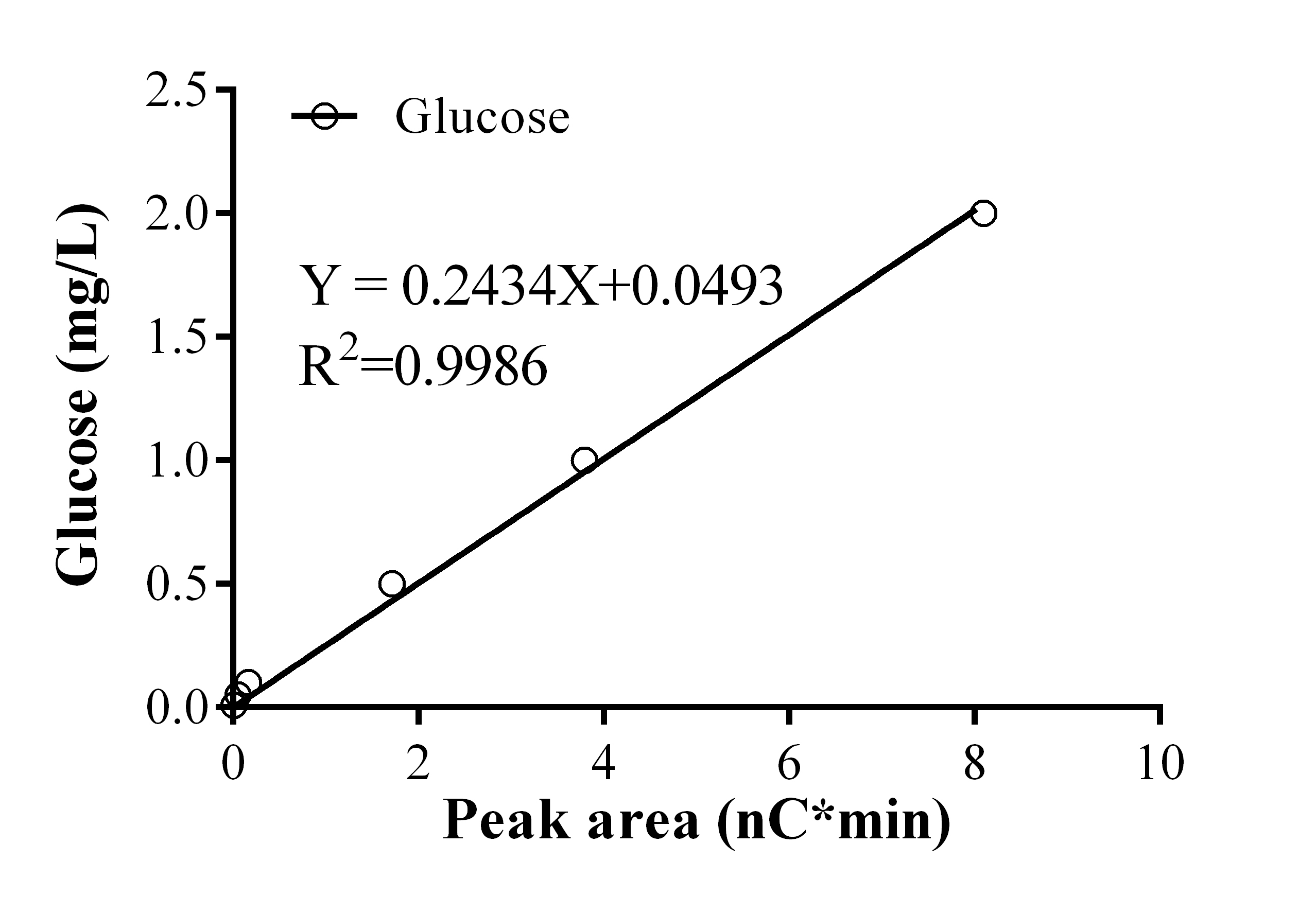
Figure 2. The glucose standard curve
- Retention times of glucose and fructose using the method described above are 3.5 and 3.8 min respectively (Figure 1).
Data analysis
Calculate the in vitro invertase enzymatic activities according to equations:
Invertase activities [nmol glucose/min/mg protein] = Cglucose x D/MWglucose/RT/Cprotein
where, Cglucose, the glucose resulted from the invertase reactions, unit: nmol/L;
D, dilution ratio for the cell crude extract, namely 50 in this protocol;
MWgluose, molecular weight of glucose, namely 180;
RT, reaction time, namely 90 min in this protocol;
Cprotein, the protein concentration of the cell crude extract, unit: mg/L.
Recipes
- BG11 medium (Rippka et al., 1979)
- 8 kinds of 10x stocks were prepared as follows:
Stock 1 (40 g/L K2HPO4·3H2O)
Stock 2 (75 g/L MgSO4·7H2O)
Stock 3 (36 g/L CaCl2·2H2O)
Stock 4 (6 g/L Critic acid)
Stock 5 (6 g/L Ferric ammonium citrate)
Stock 6 (1 g/L EDTA·2Na·2H2O)
Stock 7 (20 g/L Na2CO3)
Stock A5 (2.86 g/L H3BO3, 1.81 g/L MnCl2·4H2O, 0.22 g/L ZnSO4·7H2O, 0.39 g/L NaMoO4·2H2O, 0.08 mg/L CuSO4·5H2O, 0.01 g/L CoCl2·6H2O)
Autoclave Stocks 1 and 5 at 121 °C for 20 min. Store the autoclaved stocks and other stocks at 4 °C before use - Dissovle 1.5 g NaNO3 in 992 ml of ddH2O, and add 1 ml of Stock 2, 3, 4, 6, 7 and A5 into the medium. Autoclave the medium at 121 °C for 20 min, and supplement with 1 ml of Stocks 1 and 5
- 8 kinds of 10x stocks were prepared as follows:
- 100 mM potassium phosphate buffer (pH 7.0)
- Prepare 0.2 M K2HPO4 solution by dissolving 45.6 g K2HPO4∙3H2O in ddH2O and adjusting the volume to 1 L
- Prepare 0.2 M KH2PO4 solution by dissolving 27.2 g KH2PO4 in ddH2O and adjusting the volume to 1 L
- Prepare 100 mM potassium phosphate buffer (pH 7.0) by mixing 38 ml of 0.2 M KH2PO4 solution, 62 ml of 0.2 M KH2PO4 solution and 100 ml of ddH2O
- Prepare 0.2 M K2HPO4 solution by dissolving 45.6 g K2HPO4∙3H2O in ddH2O and adjusting the volume to 1 L
- 100 mM PMSF
Dissolve 0.174 g PMSF into 10 ml of isopropanol, and store at -20 °C before use
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars of China (31525002 to X. Lu), Shandong Key basic Research project (ZR2017ZB0211), the Joint Sino-German Research Project (grant GZ 984 to X. Lu), the Shandong Taishan Scholarship (X. Lu), the National Science Foundation of China (31301018 to X. Tan), the Key Research Program of the Chinese Academy of Sciences (ZDRW-ZS-2016-3 to X. Tan) and Qingdao Innovative Leading Talent (15-10-3-15-(31)-zch).
References
- Curatti, L., Giarrocco, L. E., Cumino, A. C. and Salerno, G. L. (2008). Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta 228(4): 617-625.
- Du, W., Liang, F., Duan, Y., Tan, X. and Lu, X. (2013). Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab Eng 19: 17-25.
- Kolman, M. A., Nishi, C. N., Perez-Cenci, M. and Salerno, G. L. (2015). Sucrose in cyanobacteria: from a salt-response molecule to play a key role in nitrogen fixation. Life (Basel) 5(1): 102-126.
- Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111(1):1-61.
- Tan, X., Yao, L., Gao, Q., Wang, W., Qi, F. and Lu, X. (2011). Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng 13(2): 169-176.
- Tan, X., Du, W. and Lu, X. (2015). Photosynthetic and extracellular production of glucosylglycerol by genetically engineered and gel-encapsulated cyanobacteria. Appl Microbiol Biotechnol 99(5): 2147-2154.
- Vargas, W., Cumino, A. and Salerno, G. L. (2003). Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 216(6): 951-960.
- Vargas, W. A. and Salerno, G. L. (2010). The Cinderella story of sucrose hydrolysis: Alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Sci 178(1):1-8.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tan, X., Song, K. and Lu, X. (2018). Enzymatic Activity Assay for Invertase in Synechocystis Cells. Bio-protocol 8(10): e2856. DOI: 10.21769/BioProtoc.2856.
Category
Microbiology > Microbial biochemistry > Carbohydrate
Microbiology > Microbial metabolism > Carbohydrate
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









