- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Explant Cultures to Interrogate Signaling Pathways that Regulate Mouse Lung Development
(*contributed equally to this work) Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2852 Views: 7048
Reviewed by: Giusy TornilloZhenguang ZhangGunjan Mehta

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2627 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1323 Views

Examining the Roles of m6A Sites in mRNA Using the Luciferase Gene Fused With Mutated RRACH Motifs
Nobuko Katoku-Kikyo and Nobuaki Kikyo
Nov 5, 2025 1938 Views
Abstract
Early mouse lung development, including specification of primordia, patterning of early endoderm and determination of regional progenitor cell fates, is tightly regulated. The ability to culture explanted embryonic lung tissue provides a tractable model to study cellular interactions and paracrine factors that regulate these processes. We provide up-to-date protocols for the establishment of this culture model and its application to investigate hedgehog signaling in the developing lung.
Keywords: Mouse embryonic lungBackground
Mouse lung development initiates as an endodermal diverticulum of the anterior foregut endoderm at day 9.5 postcoitum (E9.5), with subsequent closure of a proximal tracheoesophageal septum for the formation of distinct tracheal and esophageal tubes (Minoo and King, 1994). Subsequent branching of primitive endodermal tubes yields a planar lung structure by E12.5, with subsequent orthogonal branches yielding three-dimensional structure characteristic of the mature lung (Metzger et al., 2008). The planar structure of lung rudiments isolated prior to E12.5 is suitable for in vitro culture at an air liquid interface (Carraro et al., 2010; Del Moral and Warburton, 2010). Embryonic lung is isolated by dissection using a stereo microscope either under bright field illumination or by fluorescence illumination when coupled with lineage tracing and fluorescent reporters. Herein we describe the use of a ShhCre/RosamTmG reporter mouse allowing Cre-mediated activation of membrane-localized GFP within anterior foregut endoderm from approximately E8.75 (Montgomery et al., 2007; Goss et al., 2009; Yao et al., 2017). Accordingly lung endoderm is visualized by green fluorescence and surrounding tissue by red fluorescence, allowing clear identification and microdissection of developing endodermal structures, including the lung, and imaging during in vitro culture.
Materials and Reagents
- Whole embryonic lung isolation
- BD 1 ml TB syringe 26 G (BD, catalog number: 309625 )
- 50 ml conical tube (Denville Scientific, catalog number: C1062-P (1000799))
- Petri dish (Greiner Bio One International, catalog number: 663161 )
- ShhCre mice (THE JACKSON LABORATORY, catalog number: 005622 )
- RosamTmG/+ mice (THE JACKSON LABORATORY, catalog number: 007576 )
- Mouse embryonic lungs from ShhCre/+, RosamTmG/+ mice
Note: Mouse embryonic lungs from ShhCre/+; RosamTmG/+ mice were harvested between E10.5 and E12.5. The day of vaginal plug detection was considered to be E0.5 - General anesthesia: Ketamine (VET one, NDC 13985-702-10) and xylazine (AnaSed Injection, NDC 59339-110-20)
- 70% ethanol (Fisher Scientific, catalog number: BP8201500 )
- Phosphate buffered saline (PBS) (1x), liquid, without calcium and magnesium (Corning, catalog number: 21-040-CV )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15070063 )
- PBS with P/S (see Recipes)
- BD 1 ml TB syringe 26 G (BD, catalog number: 309625 )
- Whole embryonic lung culture
- 12 wells plate (Denville Scientific, catalog number: T1012 )
- Disposable transfer pipettes, sterile (VWR, catalog number: 414004-036 )
- Razor blade (VWR, catalog number: 55411-050 )
- Nuclepore Polycarbonate Track-Etch membrane (13 mm, 8 μm) (GE Healthcare, catalog number: 150446 )
- Dulbecco’s modified Eagle medium: Nutrient Mix F-12 (DMEM/F12) (1x), liquid, 1:1 Contains GlutaMAX, but no HEPES buffer (Thermo Fisher Scientific, GibcoTM, catalog number: 10565042 )
- Penicillin-streptomycin (P/S) (Thermo Fisher Scientific, GibcoTM, catalog number: 15070063 )
- BenchMark fetal bovine serum (Gemini Bio-Products, catalog number: 100-106 )
- Embryonic lung culture medium (see Recipes)
- 12 wells plate (Denville Scientific, catalog number: T1012 )
Equipment
- Surgical instruments including:
- Mouse necropsy instrument set including:
- Metzenbaum (Lahey) Scissors (Roboz Surgical Instrument, catalog number: RS-6950 )
- Micro Dissecting Scissors 4" Blunt (Roboz Surgical Instrument, catalog number: RS-5980 )
- Graefe Forceps Straight (Roboz Surgical Instrument, catalog number: RS-5130 )
- Graefe Forceps Curved (Roboz Surgical Instrument, catalog number: RS- 5135 )
- Metzenbaum (Lahey) Scissors (Roboz Surgical Instrument, catalog number: RS-6950 )
- CO2 Incubator (Thermo Fisher Scientific, model: HeracellTM 150i )
- Fluorescent Stereo Microscope (Carl Zeiss, model: Zeiss SteREO Discovery.V8 , with 8x magnification) equipped with 5 MP, 36 bit, Peltier cooled Zeiss Axiocam MRc5 camera (Carl Zeiss, model: AxioCam MRc 5 )
Software
- Zen blue software (Carl Zeiss)
- PRISM software version 7
Procedure
- Embryonic lung isolation
- Timed-pregnant mice (11.5-12.5 days post-coitum) are placed under general anesthesia by intraperitoneal injection of ketamine/xylazine (ketamine 100 mg/kg, xylazine 10 mg/kg) with 30 G needle according to the Institutional Animal Care and Use Committee-approved protocol (see Note 1).
- Once in a surgical plane (determined by lack of toe pinch and eye blink reflexes) the abdomen is rinsed with 70% ethanol (see Note 2).
- To collect uterus, grasp the skin with Graefe forceps and make an incision with Lahey scissors and manually open the incision. Open the peritoneal cavity, externalize the uterine structures containing embryo’s and surgically remove both uterine horns.
- Transfer the uterus to a 50 ml conical tube filled with cold PBS supplemented with P/S and rinse to remove blood.
- Transfer the uterus to a Petri dish filled with PBS supplemented with P/S and visualize using a stereo microscope with bright field fiber-optic illumination. Remove embryos from the uterus by manually opening the uterine wall with a pair of Dumont forceps. Collect and transfer the embryos with a perforated spoon to a new Petri dish kept on ice.
- Switch the microscope to the fluorescent mode and use GFP channel; from now on the dissection will be under fluorescent channels.
- Place each embryo on its left flank and visualize by epifluorescence using a 488 nm (GFP) filter (Figure 1A). Brain and limbs will be visible by auto-fluorescence. Use one pair of forceps to stabilize the embryo while gently removing skin tissue with a second pair of forceps. Remove the right forelimb first, then open the thorax along the right flank from the neck to abdomen (Figure 1B). The exposed lung and esophagus will show clear GFP fluorescence that is visible above background auto-fluorescence. The heart and the liver are removed to fully expose the lung and GI tract, all of which will are marked by GFP (Figures 1C-1D). Stomach and distal GI structures are removed and remaining GFP-positive tissue (lung and esophagus) removed using forceps to gently separate from surrounding connective tissue. Any remaining adventitial tissue should be carefully removed from the isolated lung (Figures 1E and 1F).
- Isolated lung tissue is transferred to individual wells of a 12-well plate containing embryonic lung culture medium with a sterile transfer pipet.
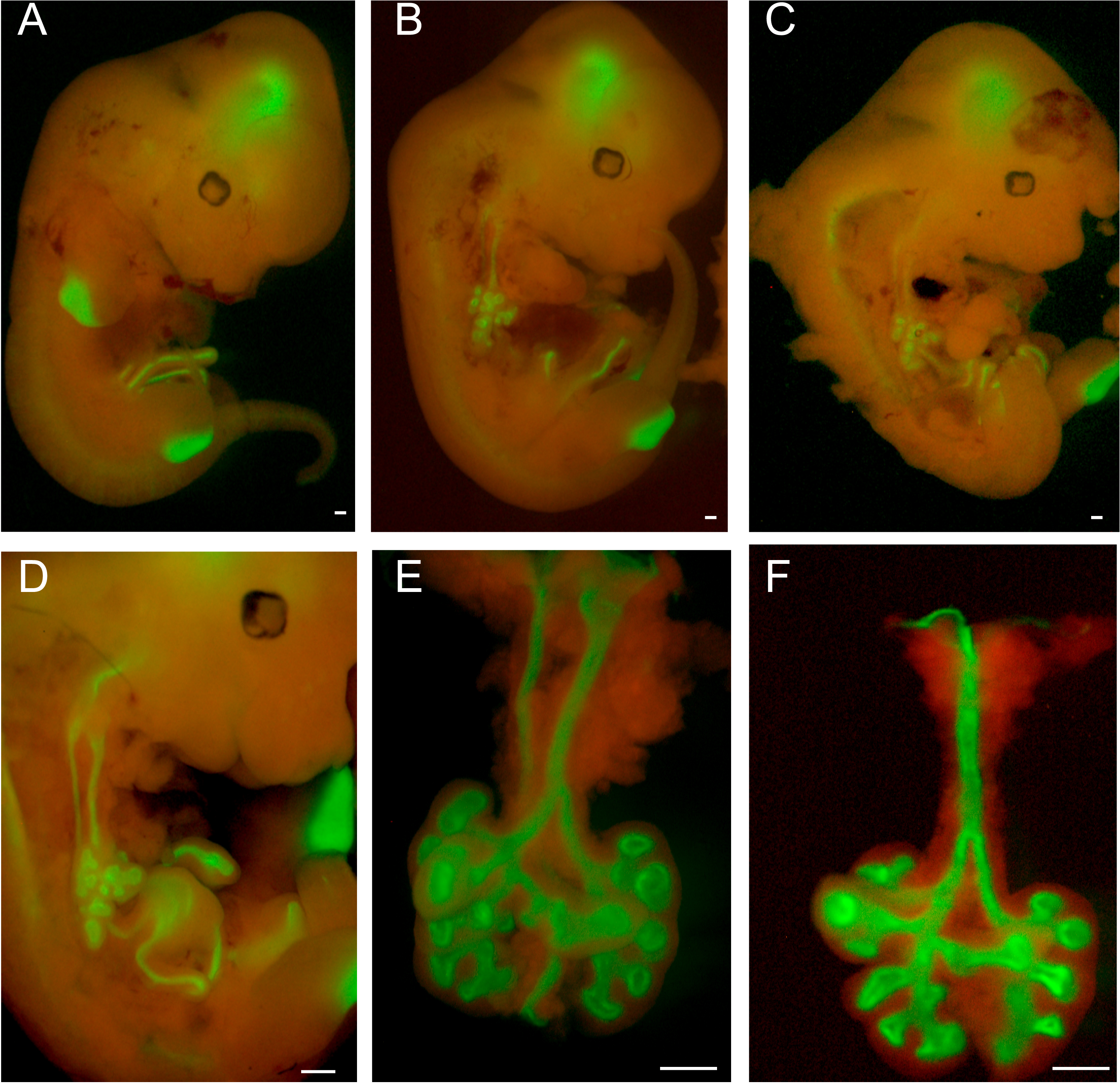
Figure 1. Dissection of E12.5 embryonic lungs from ShhCre/+; RosamTmG/+ mice by fluorescence microscopy. A. Whole embryo view of E12.5 ShhCre/+; RosamTmG/+ mice with dual detection of GFP (green) and RFP (red) fluorescence. B. View after removing the right forelimb and exposure of the thoracic cavity along the right flank. C-D. View of lung and GI tract after removal of heart and liver. E. Lung and esophagus. F. Lung after removal of the esophagus. Scale bars = 500 µm.
- Timed-pregnant mice (11.5-12.5 days post-coitum) are placed under general anesthesia by intraperitoneal injection of ketamine/xylazine (ketamine 100 mg/kg, xylazine 10 mg/kg) with 30 G needle according to the Institutional Animal Care and Use Committee-approved protocol (see Note 1).
- Embryonic lung culture
Note: From now on, every step should ideally be performed in a sterile environment such as a tissue culture hood.- Add 2 ml of embryonic lung culture medium per well.
- Place a nuclepore polycarbonate (smooth side down) track-etch membrane suspended above media in each well (see Note 3).
- Use a sharp sterile razor blade to remove approximately 1.5 cm of a 200 μl pipet tip and use the resulting wide bore to gently capture one embryonic lung and transfer to the upper surface of the nuclepore polycarbonate track-etch membrane.
- Ensure that the lung is flat on the polycarbonate membrane by visualization under an inverted light microscope; gently adjust using fine forceps to ensure that it is flat.
- Image the lung as day 0 (Figure 2).
- Any treatment, including inhibitors, growth factors, siRNA etc., should be included in culture media at this time.
- Transfer the 12-well plate to a humidified cell culture incubator at 37 °C and monitor the experiment by imaging at the desired time points (Figure 2).
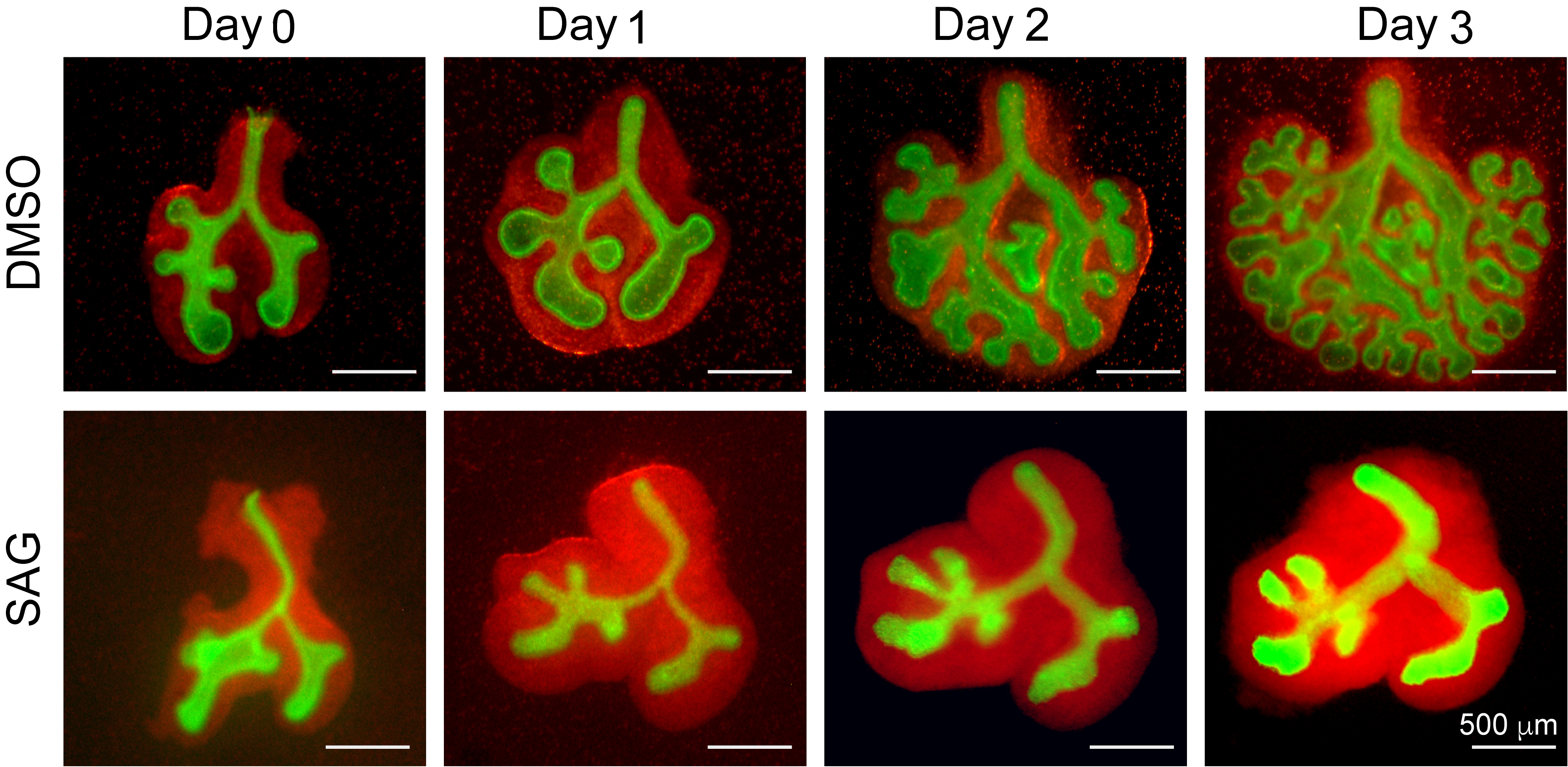
Figure 2. Potentiation of the hedgehog pathway by culturing in the presence of the smoothened receptor agonist SAG, induces mesodermal proliferation and blocks endoderm branching. Panels show whole-mount fluorescence images of embryonic lung endoderm (green) and mesoderm (red) cultured for 0-3 days in the presence of media containing DMSO vehicle (upper 4 panels) or media supplemented with 10 µM SAG (lower 4 panels). SAG treatment induces loss of branching and abnormal growth of mesodermal cells (red) indicative of roles played by Sonic Hedgehog signaling in the appropriate regulation of lung branching.
- Add 2 ml of embryonic lung culture medium per well.
Data analysis
Images were taken and processed using Zen blue software, statistics analysis was performed using PRISM software version 7.
Notes
- Any animal work should follow federal and local regulation.
- To check how deep is the anesthetization, use a pair of forceps to pinch toes and check if there is any reflection, do not proceed until there is totally no reflection.
- Make sure the shiny smooth side of the nuclepore polycarbonate track-etch membrane face down towards the media and the rough side face up. The lungs would be transferred with a drop of media surrounded on top of the rough side without slipping away.
Recipes
- PBS with P/S
1x phosphate buffered saline (PBS), liquid, without calcium and magnesium
50 U/ml of penicillin-streptomycin - Embryonic lung culture medium
DMEM/F-12, GlutaMAXTM (Nutrient Mix F-12 (1x), liquid, 1:1, Contains GlutaMAX, but no HEPES buffer)
50 U/ml of penicillin-streptomycin (P/S)
10% BenchMark fetal bovine serum
Acknowledgments
This work was funded by the National Institutes of Health 1T32HL134637-01, R01HL135163-01; California Institution of Regenerative Medicine CIRM LA1-06915. The protocol described here is adapted from Yao et al. (2017). All authors declared that no competing interest exists.
References
- Carraro, G., del Moral, P. M. and Warburton, D. (2010). Mouse embryonic lung culture, a system to evaluate the molecular mechanisms of branching. J Vis Exp (40).
- Del Moral, P. M. and Warburton, D. (2010). Explant culture of mouse embryonic whole lung, isolated epithelium, or mesenchyme under chemically defined conditions as a system to evaluate the molecular mechanism of branching morphogenesis and cellular differentiation. Methods Mol Biol 633: 71-79.
- Goss, A. M., Tian, Y., Tsukiyama, T., Cohen, E. D., Zhou, D., Lu, M. M., Yamaguchi, T. P. and Morrisey, E. E. (2009). Wnt2/2b and β-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell 17(2): 290-298.
- Metzger, R. J., Klein, O. D., Martin, G. R. and Krasnow, M. A. (2008). The branching programme of mouse lung development. Nature 453(7196): 745-750.
- Minoo, P. and King, R. J. (1994). Epithelial-mesenchymal interactions in lung development. Annu Rev Physiol 56: 13-45.
- Montgomery, R. L., Davis, C. A., Potthoff, M. J., Haberland, M., Fielitz, J., Qi, X., Hill, J. A., Richardson, J. A. and Olson, E. N. (2007). Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21(14): 1790-1802.
- Yao, C., Carraro, G., Konda, B., Guan, X., Mizuno, T., Chiba, N., Kostelny, M., Kurkciyan, A., David, G., McQualter, J. L. and Stripp, B. R. (2017). Sin3a regulates epithelial progenitor cell fate during lung development. Development 144(14): 2618-2628.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yao, C., Carraro, G. and Stripp, B. R. (2018). In vitro Explant Cultures to Interrogate Signaling Pathways that Regulate Mouse Lung Development. Bio-protocol 8(10): e2852. DOI: 10.21769/BioProtoc.2852.
Category
Developmental Biology > Morphogenesis > Organogenesis
Cell Biology > Tissue analysis > Tissue isolation
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








