- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Hydrogen Sulfide and Cysteine Excreted by Bacterial Cells
Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2847 Views: 7131
Reviewed by: Valentine V TrotterDarrell CockburnAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Mn Concentrations in Synechocystis sp. PCC6803 Using ICP-MS
Fabian Brandenburg [...] Marion Eisenhut
Dec 5, 2017 8588 Views

Extraction and Quantification of Polyphosphate (polyP) from Gram-negative Bacteria
Jan-Ulrik Dahl [...] Ursula Jakob
Sep 20, 2018 7045 Views

Flow Cytometry-based Measurement of Reactive Oxygen Species in Cyanobacteria
Soumila Mondal and Shailendra P. Singh
May 20, 2022 3835 Views
Abstract
Bacteria release cysteine to moderate the size of their intracellular pools. They can also evolve hydrogen sulfide, either through dissimilatory reduction of oxidized forms of sulfur or through the deliberate or inadvertent degradation of intracellular cysteine. These processes can have important consequences upon microbial communities, because excreted cysteine autoxidizes to generate hydrogen peroxide, and hydrogen sulfide is a potentially toxic species that can block aerobic respiration by inhibiting cytochrome oxidases. Lead acetate strips can be used to obtain semiquantitative data of sulfide evolution (Oguri et al., 2012). Here we describe methods that allow more-quantitative and discriminatory measures of cysteine and hydrogen sulfide release from bacterial cells. An illustrative example is provided in which Escherichia coli rapidly evolves both cysteine and sulfide upon exposure to exogenous cystine (Chonoles Imlay et al., 2015; Korshunov et al., 2016).
Keywords: Hydrogen sulfideBackground
Reduced sulfur species are generated by microbes through several routes. Sulfate-reducing bacteria exploit the reductive process as an integral part of energy generation. Other bacteria release sulfide as a by-product of either the deliberate or adventitious degradation of sulfur species, including cysteine. We have observed that cysteine itself is excreted when intracellular levels are abnormally high, a situation that can occur through uncontrolled amino acid import or dysregulation of cysteine synthesis. These sulfur species are unusually reactive, as they bind metals with high avidity and also are among the few biomolecules that react chemically with molecular oxygen. The upshot is that reduced sulfur compounds can exert important effects upon cells. Hence, it can be important to track the dynamics of reduced sulfur compounds in a variety of contexts.
Thiol agents–notably, 5,5-dithiobis (2-nitrobenzoic acid) (DTNB)–provide good spectroscopic probes of thiol concentrations. Unfortunately, they do not discriminate between organic thiols like cysteine and inorganic species like hydrogen sulfide. The evolution of the latter species has often been detected using lead acetate strips, which are suspended in the head space over sulfide-generating cultures. However, this method is slow and non-quantitative. For that reason, we have leveraged the volatility of hydrogen sulfide so that standard dyes can allow sulfide and organic thiols to be distinguished as they are generated in lab cultures. The methods are simple, quick, and sensitive.
Materials and Reagents
- 10 ml test tubes, as necessary (Fisher Scientific, catalog number: 14-961-27 )
- Polypropylene tubes, 2 ml, as necessary (Denville Scientific, catalog number: C2170 )*
- Polypropylene 2 ml tubes, as necessary (see Material and Reagents #2)**
- Parafilm* (Bemis, catalog number: PM996 )
- Pipette tips (1,000 μl; 200 μl) (Corning, catalog number: 4846 ; USA Scientific, catalog number: 1111-1006 )
- Cylinder with compressed air (AirGas Mid-America, breathing quality grade D)
- Cylinder with compressed nitrogen (AirGas Mid-America)**
- 50 ml flasks (Corning, PYREX®, catalog number: 4442-50 )
- Flask closures (Fisher Scientific, catalog number: 05-888 )
- Bacterial cell culture
- Ethylenediamine tetraacetic acid, disodium salt, dihydrate, EDTA (Fisher Scientific, catalog number: S311 )
- Cystine dihydrochloride (Sigma-Aldrich, catalog number: C2526 )
- 5,5-Dithiobis (2-nitrobenzoic acid), DTNB (Sigma-Aldrich, catalog number: D8130 )
- 4-Amino-N,N-dimethylaniline, DMPDA (Sigma-Aldrich, catalog number: 07750 )
- Ferric(III) chloride hexahydrate (Sigma-Aldrich, catalog number: F2877 )
- Potassium phosphate, mono- or dibasic (Fisher Scientific, catalog numbers: P284 , P288 )
- Ethanol, 100% (Decon Labs, catalog number: 2716 )
- Potassium hydroxide (Fisher Scientific, catalog number: P250 )
- Glucose (Fisher Scientific, catalog number: D16 )
- Ammonium sulfate (Fisher Scientific, catalog number: A702 )
- Sodium citrate (Fisher Scientific, catalog number: S279 )
- Hydrochloric acid (Sigma-Aldrich, catalog number: H1758 )
- Sodium sulfide nonahydrate (Sigma-Aldrich, catalog number: S4766 )
- Magnesium sulfate heptahydrate (Fisher Scientific, catalog number: M63 )
- Deionized water (University of Illinois deionizing system)
- Stock solutions (see Recipes)
*These items are for DMPDA-based measurements.
**These items are for DTNB-based measurements.
Equipment
- For DMPDA-based measurements
- Pipettors, 1 and 0.2 ml (Mettler-Toledo, Rainin, catalog numbers: 17014382 , 17014384 )
- Microcentrifuge (Fisher Scientific, model: accuSpinTM Micro 17 , catalog number: 13-100-675)
- Spectrophotometer (Beckman Coulter, model: DU-640 )
- Shaking water bath (New Brunswick Scientific, model: G76D )
- Heater (Fisher Scientific, catalog number: 11-718 )
- Pipettors, 1 and 0.2 ml (Mettler-Toledo, Rainin, catalog numbers: 17014382 , 17014384 )
- For DTNB-based measurements
- Two 125 ml gas washing bottles with coarse fritted discs (Corning, PYREX®, catalog number: 31760-125C )
- Water bath (Shel-Lab, VWR, model: Model 1250 )
- Two 125 ml gas washing bottles with coarse fritted discs (Corning, PYREX®, catalog number: 31760-125C )
Procedure
- Measurement of excreted organic thiols
- Grow E. coli cultures (> 5 ml) to OD 0.1 in a medium that contains sulfate as the sole sulfur source, such as minimal A medium (Chonoles Imlay et al., 2015).
- Add 0.1 mM EDTA to the bacterial culture.
- Add 0.2 mM cystine to the culture. Turn the shaker on and let the culture mix for 10 sec.
- Remove 1 ml of bacterial culture every 2-5 min.
- Centrifuge the 1-ml aliquot (30-60 sec) using microcentrifuge (13,800 x g).
- Place 0.5 ml of supernatant into a test tube and bubble it vigorously with nitrogen for minimum 30 sec to remove hydrogen sulfide.
- Mix the bubbled supernatant with 0.5 ml of DTNB reagent by pipetting 3-4 times.
- Allow reaction for minimum 1 min and then measure the absorbance at 412 nm.
- Calculate the thiol concentration using extinction coefficient of 13 OD/mM of cysteine. Multiply the obtained value by 2 to correct for the dilution by the DTNB reagent.
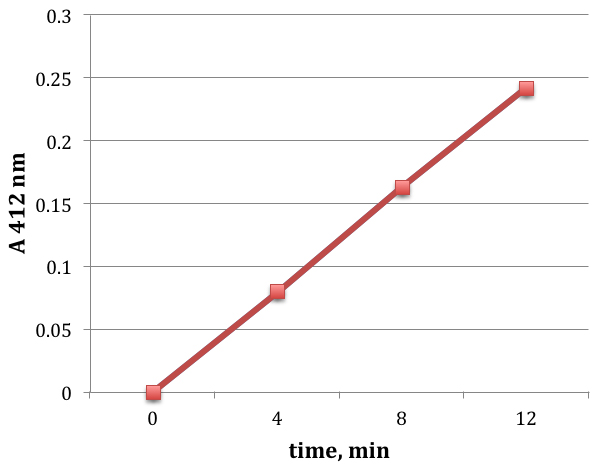
Figure 1. Sample time course of cysteine excretion. The progressive accumulation of cysteine in the supernatant of wild-type (MG1655) E. coli cultures was tracked after cystine addition. Cells were at 0.1 OD in minimal glucose medium. Cystine was added at time point zero. The rate of cysteine accumulation in the medium is 3.1 μM/min. - Grow E. coli cultures (> 5 ml) to OD 0.1 in a medium that contains sulfate as the sole sulfur source, such as minimal A medium (Chonoles Imlay et al., 2015).
- One-time sulfide measurements for sealed cultures
- Grow E. coli cultures to OD 0.1 in a medium that contains sulfate as the sole sulfur source, such as minimal A medium (Chonoles Imlay et al., 2015).
- Transfer 1 ml of bacterial culture into a 2 ml plastic tube in a heater at 37 °C.
- Add 0.1 mM cystine. Close the tube and seal it with Parafilm.
- Incubate for 10 min at 37 °C. Agitation is not necessary.
- Remove Parafilm, open the tube and quickly add 0.1 ml of 20 mM DMPDA followed immediately by 0.1 ml of 30 mM ferric chloride. Acidification will lyse cells; the DMPDA/ferric chloride will detect sulfide that had been released during the prior incubation period. Close the tube and seal it with Parafilm. To avoid the loss of hydrogen sulfide, do not pipet or mix the culture until the tube is closed and sealed.
- Shake the sealed tube and keep it in the dark at room temperature for 15 min. The control culture should not contain cystine.
- After the incubation, remove Parafilm and centrifuge the samples in a table centrifuge for 5 min at 13,800 x g.
- Carefully transfer 1 ml of the supernatant to clean plastic tubes.
- Measure the optical density at 650 nm.
- The extinction coefficient may vary depending on the medium and lies between 16-20 OD/mM of sulfide with typical mean of 17 OD/mM. In some media (for example, glycerol-containing minimal A medium) the absorbance value of the sample may drift over the time of incubation with DMPDA; therefore, the time span between the addition of DMPDA and the measurement of absorbance should be as consistent as possible and should not differ between samples by more than 2-3 min. A calibration curve using sodium sulfide in the 0.5-20 micromolar range should be obtained for each medium. This method was modified from Siegel (1965). Typical results are presented in Figure 2.
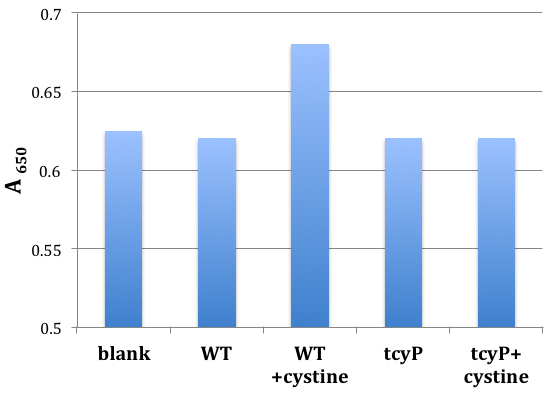
Figure 2. Sample single-point sulfide measurements in bacterial cultures. A wild-type MG1655 cell culture (OD 0.1) in minimal A glucose medium was incubated for 10 min in the absence or presence of 0.2 mM of cystine, and the evolved hydrogen sulfide was then measured using DMPDA. Sterile medium was used as a blank; note the substantial background value. The suspension from cystine-fed wild-type cells accumulated 3.5 micromolar sulfide. The strain that was unable to import cystine into the cell (tcyP) did not produce any detectable sulfide.
- Grow E. coli cultures to OD 0.1 in a medium that contains sulfate as the sole sulfur source, such as minimal A medium (Chonoles Imlay et al., 2015).
- Time course for sulfide accumulation in open growing cultures
- Grow E. coli cultures to OD 0.1 in minimal A medium, with sulfate as the sole sulfur source.
- Place 12 ml of the culture into a 50 ml flask.
- Add 0.2 mM of cystine.
- Seal the flask with stopper. Shake the culture at 180 rpm.
- Take 1 ml samples every 5 min. Do not keep the flask open for more than 5 sec.
- Perform Steps B4-B8 from ‘one-time measurements for sealed cultures’ section.
The DMPDA-based method for sulfide determination may not work properly if high cysteine concentrations (> 5 mM) are present in the medium. The weakness of this method is that, despite high reliability and sensitivity, the signal/background ratio is not optimal at low sulfide concentrations (see Figure 1). To address this situation, we developed a simple and sensitive DTNB-dependent system that employs an alkaline trap. - Grow E. coli cultures to OD 0.1 in minimal A medium, with sulfate as the sole sulfur source.
- DTNB-based procedure with an alkaline trap
- Grow E. coli culture to OD 0.1 in minimal A medium, with sulfate as the sole sulfur source.
- Connect the outlet of one gas-washing bottle with the fritted tube of the other (Figure 3).
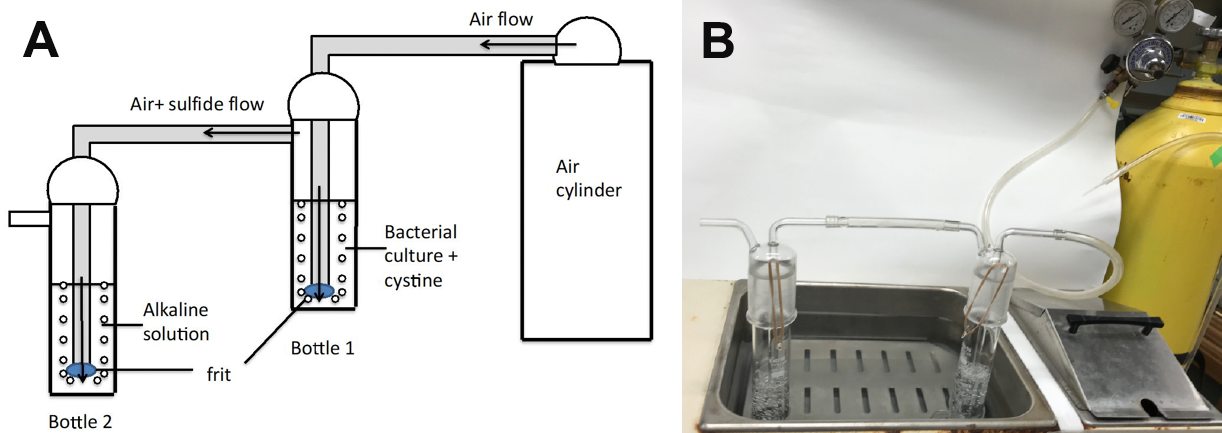
Figure 3. Arrangement for continuous trapping of sulfide released by cultures. A. Bottle 1 contains the active bacterial culture and may be located in a water bath. The inlet of the bottle 1 is connected to the cylinder with compressed air; the outlet of the bottle is connected to the inlet of the bottle 2 containing the alkaline solution. The outlet of the bottle 2 is not sealed. After addition of cystine to the bottle 1 the cap of this bottle is fixed by the rubber or metallic spring hooks to the bottle’s body. At intervals, samples from bottle 2 and removed, and sulfide is assayed. B. Photographic depiction of the sulfide trap. - Fill the first bottle (bottle 1) with 75-100 ml of the bacterial culture (0.3-0.4 OD).
- Fill another bottle (bottle 2) with an equal volume of 0.1 M KPO4 buffer at pH 11.
- Set the air flow to 0.3-0.4 L/min. (Air flow is easily measured by quantifying the rate of water displacement when the line is directed into an inverted water-filled graduated cylinder.)
- Add 0.5 mM of cystine and seal the bottle. The flow of air passes through the bacterial culture and carries the released hydrogen sulfide to the alkaline solution in the next bottle. Hydrogen sulfide is deprotonated and thereby trapped in the alkaline solution of bottle 2.
- Periodically remove 0.5 ml of the alkaline solution and mix it with 0.5 ml of 0.5 mM DTNB solution in 0.5 M KPO4 (pH 7). Determine absorbance at 412 nm.
- Calculate hydrogen sulfide content using extinction coefficient of 26 OD/mM of hydrogen sulfide. Note that a correction should be made for the 1:1 dilution of the sample solution into the DTNB solution.
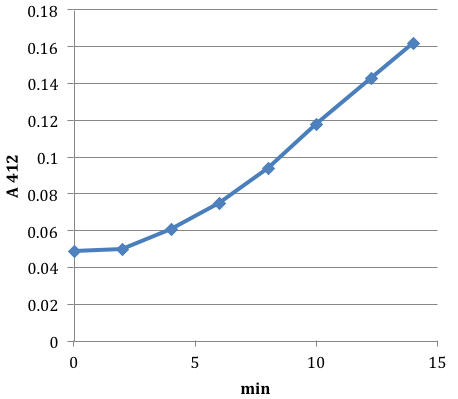
Figure 4. Sample time course of sulfide accumulation in an alkaline trap. Hydrogen sulfide formation was initiated by the addition of 0.5 mM cystine to an MG1655 culture (0.3 OD) in minimal A glucose medium. After addition of cystine the cell culture was sealed. Air flow rate was 0.35 l/min, the suspension volume was 75 ml, the trap volume was 75 ml. 0.1 KPO4 buffer taken as a blank. At intervals, aliquots were removed from the trap and thiol content was quantified by DTNB treatment. The initial lag represents the time needed for initial sulfide accumulation in the culture; the final slope represents the rate of sulfide evolution by the bacteria. - Grow E. coli culture to OD 0.1 in minimal A medium, with sulfate as the sole sulfur source.
- Determination of the efficiency of the alkaline trap
- Fill bottle 1 with 75-100 ml of the medium containing 10 micromolar sodium sulfide.
- Fill bottle 2 with an equal volume of alkaline solution.
- Take 0.5 ml samples from each bottle and mix with 0.5 ml of the DTNB solution.
- Seal bottle 1 and adjust the air flow to 0.3-0.4 L/min.
- Bubble air for 10 min and repeat Step E3.
- Compare the change in OD from both bottles. The drop in OD from the pre- and post-gassing samples of bottle 1 represents the amount of sulfide that was removed by gassing. The rise in OD from the pre- and post-gassing samples of bottle 2 represents the amount of sulfide that was successfully trapped. Thus the efficiency of the trap is (ΔOD bottle 2)/(ΔOD bottle 1). The efficiency observed in our experiments was about 80-85%.
- Fill bottle 1 with 75-100 ml of the medium containing 10 micromolar sodium sulfide.
Data analysis
Cysteine-excretion and sulfide-excretion rates are calculated from the slope of the data (see Figures 1 and 3), using the absorbance values of standard samples to convert rate of absorbance change to rate of analyte concentration change. DMPDA determinations of sulfide in bacterial cultures rely on single measurements per sample; blanks from sulfide-free samples are subtracted before calculating the sulfide content of experimental samples. Technical replicates can be determined for each point and averaged, and best-fit curves are calculated. As discussed below, most variation arises from biological variance, which has different sources depending upon the phenomenon being studied; accordingly, workers are encouraged to measure biological replicates, but the nature and degree of the variation are not addressed here.
The technical variation among samples is modest. Three micromolar cysteine in culture supernatants was measured with a standard error of 2%. Three micromolar sulfide in culture supernatants was detected with SEM of 7%. Three micromolar sulfide in an alkaline trap was detected with SEM of 2%. Precision in the cysteine and sulfide-trap analyses is improved by obtaining multiple time points, if the experimenter is confident that the actual rates of excretion do not change over the course of the measurement.
Notes
Variability in data is dominated by variance among biological rather than technical replicates. There are a variety of scenarios that might attract the interest of biologists, and so we do not attempt to highlight the issues that might impinge upon fluctuations in cell behavior. In general, the excretion of hydrogen sulfide depends upon the efficiencies of cysteine/cystine import and the titers of desulfidases; each of the responsible proteins is regulated (Korshunov et al., 2016). Similarly, cysteine efflux rates also depend upon the induced levels of export systems (Chonoles Imlay et al., 2015). Therefore, experimenters who wish to track rates of sulfide or cysteine efflux are encouraged to tightly control cell-handling protocols, particularly with respect to medium, time, and temperature.
Recipes
- Stock solutions (prepared freshly, stored on ice)
50 mM DTNB in 100% ethanol
20 mM DMPDA in 6 M hydrochloric acid
30 mM ferric chloride in 1 M hydrochloric acid
50 mM cystine in 0.2 M hydrochloric acid
50 mM EDTA, pH 8.0 - Buffers and medium stocks for the medium
0.5 M phosphate buffer: 5 g KH2PO4, 11 g K2HPO4, 200 ml of deionized water; adjust pH to 7.0
0.1 M phosphate buffer for the alkaline trap: 20 ml of 0.5 M phosphate buffer mixed with 80 ml of deionized water. Adjust pH to 11 using 6 M KOH
0.5 M magnesium sulfate in deionized water, autoclave for 30 min
20% glucose (w/v): 20 g glucose in 100 ml of deionized water, autoclave for 30 min - Minimal A medium
1.05 g K2HPO4
0.45 g KH2PO4
0.1 g (NH4)2SO4
0.05 g sodium citrate dihydrate
100 ml of deionized water
Note: Autoclave the medium for 30 min, cool to room temperature, and then add 0.1 ml of 0.5 M magnesium sulfate and 1 ml of 20% glucose.
Acknowledgments
This work was supported by grant GM101012 from the National Institutes of Health. The DMPDA-based method was previously described in Siegel (1965), Anal Biochem 11: 126-132.
There are no conflicts of interest or competing interests to be declared.
References
- Chonoles Imlay, K. R., Korshunov, S. and Imlay, J. A. (2015). Physiological roles and adverse effects of the two cystine importers of Escherichia coli. J Bacteriol 197(23): 3629-3644.
- Korshunov, S., Imlay, K. R. and Imlay, J. A. (2016). The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol Microbiol 101(1): 62-77.
- Oguri, T., Schneider, B. and Reitzer, L. (2012). Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J Bacteriol 194(16): 4366-4376.
- Siegel, L. M. (1965). A direct microdetermination for sulfide. Anal Biochem 11: 126-132.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Korshunov, S. and Imlay, J. A. (2018). Quantification of Hydrogen Sulfide and Cysteine Excreted by Bacterial Cells. Bio-protocol 8(10): e2847. DOI: 10.21769/BioProtoc.2847.
Category
Microbiology > Microbial physiology > Stress response
Microbiology > Microbial metabolism > Nutrient transport
Biochemistry > Other compound > Thiol
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









