- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detecting the Interaction of Double-stranded RNA Binding Protein, Viral Protein and Primary miRNA Transcript by Co-immunoprecipitation in planta
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2840 Views: 7049
Reviewed by: Arsalan DaudiAswad KhadilkarAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3033 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2019 Views
Abstract
MicroRNAs (miRNAs) play important roles in plant growth, development, and response to infection by microbes. Double-stranded RNA binding protein 1 (DRB1) facilitates the processing of primary miRNA transcripts into mature miRNAs. Recently, we found that NS3 protein encoded by rice stripe virus (RSV) associates with DRB1 and promotes miRNA biogenesis during RSV infection (Zheng et al., 2017). RNA co-immunoprecipitation (RIP) method was applied to identity association patterns among DRB1, NS3, and miRNA transcript.
Keywords: Rice stripe virusBackground
MicroRNAs (miRNAs) are processed from their primary transcripts (pri-miRNAs) by the RNase III enzyme DICER-LIKE 1 (DCL1) with the help of the double-stranded RNA (dsRNA) binding protein HYPONASTIC LEAVES1 (DRB1/HYL1) and the zinc finger protein SERRATE (SE). Rice stripe virus (RSV) infection broadly perturbs miRNA accumulation. We found that RSV-encoding nonstructural protein 3 (NS3) promotes miRNA accumulation by downregulating pri-miRNAs through interaction with DRB1 in rice (Zheng et al., 2017). To reveal how NS3 enhances pri-miRNA processing, we used co-immunoprecipitation (Co-IP) to illustrate the relationship of NS3, DRB1 and pri-miRNA in vivo. This protocol contributes to understand association patterns between two proteins and one RNA transcript.
Materials and Reagents
- Pipette (RNase free 1 ml, 0.2 ml and 0.02 ml, Axygen)
- Miracloth (Merck, Calbiochem, catalog number: 475855 )
- Centrifuge tube (1.5 ml, 2 ml and 50 ml) (Corning)
- 4- to 6- weeks old Nicotiana benthamiana (leaves, grow in green house)
- Agrobacterium tumefaciens (EHA105 strain, preserved in our lab)
- Expression plasmids: pEarleyGate202-DRB1, -mutant DRB1, pEarleyGate203-NS3, -mutant NS3 and pCAMBIA-artificial primary miRNA transcript, -mutant primary miRNA transcript (Constructed by ourselves)
- Double-distilled or MilliQ water (ddH2O)
- Formaldehyde (Sigma-Aldrich, catalog number: F8775 )
- PBS (Thermo Fisher Scientific, GibcoTM, catalog number: 70011069 )
- Glycine (Sigma-Aldrich, catalog number: V900144 )
- Liquid nitrogen
- Anti-Myc (9E10) monoclonal antibodies (Sigma-Aldrich, catalog numbers: M4439 ) and mouse IgG control (Thermo Fisher Scientific, Invitrogen, catalog number: 02-6502 )
- Protein G-agarose (Roche Diagnostics, catalog number: 11243233001 )
- Trizol
- Chloroform (Sigma-Aldrich, catalog number: 613304 )
- GlycoBlueTM coprecipitant (15 mg/ml) (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9516 )
- Ethanol (Sigma-Aldrich, catalog number: E7023 )
- Ethanol (Aladdin, catalog number: E111963 )
- DNase I (Promega, catalog number: M6101 )
- Sucrose (Sigma-Aldrich, catalog number: V900116 )
- Ficoll 400 (Sigma-Aldrich, catalog number: F9378 )
- Dextran T40 (Sigma-Aldrich, catalog number: 1179708 )
- HEPES (Sigma-Aldrich, catalog number: RDD002 )
- KOH
- Triton X-100 (Sigma-Aldrich, catalog number: T9284 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- EDTA-free protease inhibitor cocktail (Roche Diagnostics, catalog number: 05892953001 )
- DTT (DL-Dithiothreitol) (Sigma-Aldrich, catalog number: 43817 )
- Tris-HCl (Sigma-Aldrich, catalog number: V900312 )
- NP-40 (Sigma Aldrich, catalog number: NP40S )
- RNase inhibitor (RNaseOUT) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777019 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: V900312 )
- Diethyl pyrocarbonate (Sigma-Aldrich, catalog number: 40718 )
- Honda buffer (see Recipes)
- High salt nuclear lysis buffer (see Recipes)
- Dilution buffer (see Recipes)
- IP buffer (see Recipes)
Equipment
- Eppendorf pipettes suite (1 ml, 0.2 ml, 0.02 ml, 0.01 ml and 0.0025 ml)
- Thermomixer C (Eppendorf, model: Thermomixer® C , catalog number: 5382000023)
- Vacuum pump (FJC, model: 6912 )
- Rotator (Glas-Col, model: 099A MR1512 )
- Centrifuge (Eppendorf, models: 5424 R and 5804 R )
- Pico Ultrasonicator (Diagenode, model: Bioruptor® Pico, model: 4486126 )
- Vortex (Kylin-Bell Lab Instruments, model: VORTEX-5 )
- Pestle
Procedure
- Material preparation
Agrobacterium tumefaciens-mediated transient co-expression of corresponding proteins and RNAs was performed with 4- to 6-week old N. benthamiana. N. benthamiana leaves (~8 g). Harvest the samples at 3 dpi (days post inoculation). - Nuclei isolation
- Cross-link N. benthamiana leaves with 1% formaldehyde in 1x PBS for 15 min.
Note: Make sure the sample is submerged in the buffer, apply the vacuum for 5 min, release, Reapply vacuum, repeat this for 10 times. - Add glycine to a final concentration of 0.125 M, mix well the buffer and apply vacuum for 5 min to stop cross-linking.
- Rinse the plants three times with sterile water. Remove excess water, freeze the plants in liquid nitrogen. If not proceed to the next step immediately, store the frozen plants at -80 °C.
- Grind the plant material into fine powder with liquid nitrogen in mortar. Transfer the power to a 50 ml centrifuge tube.
- For each sample (2 g), add 15 ml Honda buffer, rotate at 4 °C for 5 min.
- Filter through one layer of Miracloth, then wash the Miracloth by adding another 10 ml of Honda buffer (in a 50 ml centrifuge tube).
- Centrifuge at 3,500 x g for 5 min at 4 °C. Discard the supernatant, softly resuspend the pellet in 1 ml Honda buffer with 1 ml pipet with end-cut tip.
- Centrifuge at 3,500 x g for 5 min, wash the pellet once more with 1 ml Honda buffer. Centrifuge at 10,000 x g for 1 min and remove the supernatant thoroughly.
- Cross-link N. benthamiana leaves with 1% formaldehyde in 1x PBS for 15 min.
- Immunoprecipitation
- Add 2.5 volumes/weight high salt nuclear lysis buffer in nuclear pellets, resuspend nuclei and sonicate (30 sec long pulses, 30 sec intervals, 10 cycles) by using a Diagenode Bioruptor.
- Centrifuge (16,000 x g, 4 °C) for 10 min, take supernatant (400 μl) into a 2 ml centrifuge tube, add four volumes of dilution buffer, mix well, take out 1/20, and keep it as input.
- Add anti-Myc (10 μg/2 ml) antibody, rotate at 4 °C for 2 h, then add 50 μl of Protein G and rotate for another 2 h.
- Centrifuge at 1,500 x g for 5 min at 4 °C. Remove supernatant carefully and add IP buffer. Wash beads with IP buffer for 3 times.
- Take out 1/10 of IP extract for Western blot detection.
- Centrifuge at 1,500 x g for 5 min at 4 °C, remove the supernatant thoroughly.
- Add 2.5 volumes/weight high salt nuclear lysis buffer in nuclear pellets, resuspend nuclei and sonicate (30 sec long pulses, 30 sec intervals, 10 cycles) by using a Diagenode Bioruptor.
- RNA extraction and RT-PCR
- Add 1 ml Trizol, 65 °C for 10 min.
- Add 200 μl chloroform, vortex, incubate the tubes on ice for 5 min.
- Centrifuge at 16,000 x g for 5 min.
- Take 400 μl aqueous phase.
- Add 1 μl GlycoBlue and 40 μl sodium acetate, then add 1 ml anhydrous ethanol, incubate at -80 °C, 2 h.
- Centrifuge at 16,000 x g for 5 min, discard the supernatant.
- Suspend the RNA pellet in 89 μl of nuclease-free water.
- Add 10 μl 10x DNase I buffer and 1 μl DNase I, 37 °C for 10 min.
- Add 300 μl nuclease-free water then 400 μl chloroform, vortex.
- Centrifuge at 16,000 x g for 5 min.
- Take 400 μl aqueous phase.
- Add 1 μl GlycoBlue and 40 μl sodium acetate, then add 1 ml anhydrous ethanol, -80 °C, 2 h.
- Centrifuge at 16,000 x g for 5 min, discard the supernatant.
- Suspend the RNA pellet in 20 μl of nuclease-free water.
- Reverse transcript by SuperScriptIII (follow the manufacturer's instructions) with gene-specific primer, and detect RNAs by RT-PCR (melting temp.: 95 °C, annealing temp.: 60 °C, cycle numbers: 30 cycles).
- Add 1 ml Trizol, 65 °C for 10 min.
Data analysis
Co-IP products were reversely transcribed using gene specific primer. Reverse transcripts were separated by electrophoresis on a 2% agarose gel. As shown in Figure 1, both OsDRB1a and mOsDRB1a interacted with apri-miR528, but neither of them recognized mapri-miR528; and NS3 and mNS3 were associated with OsDRB1a instead of mOsDRB1a. With the expression of NS3, both OsDRB1a and mOsDRB1a interacted with apri-miR528. However, with the expression of mNS3, only mOsDRB1a was associated with apri-miR528. These results indicate that NS3 may act as a scaffold to mediate the interaction between OsDRB1 and pri-miRNA.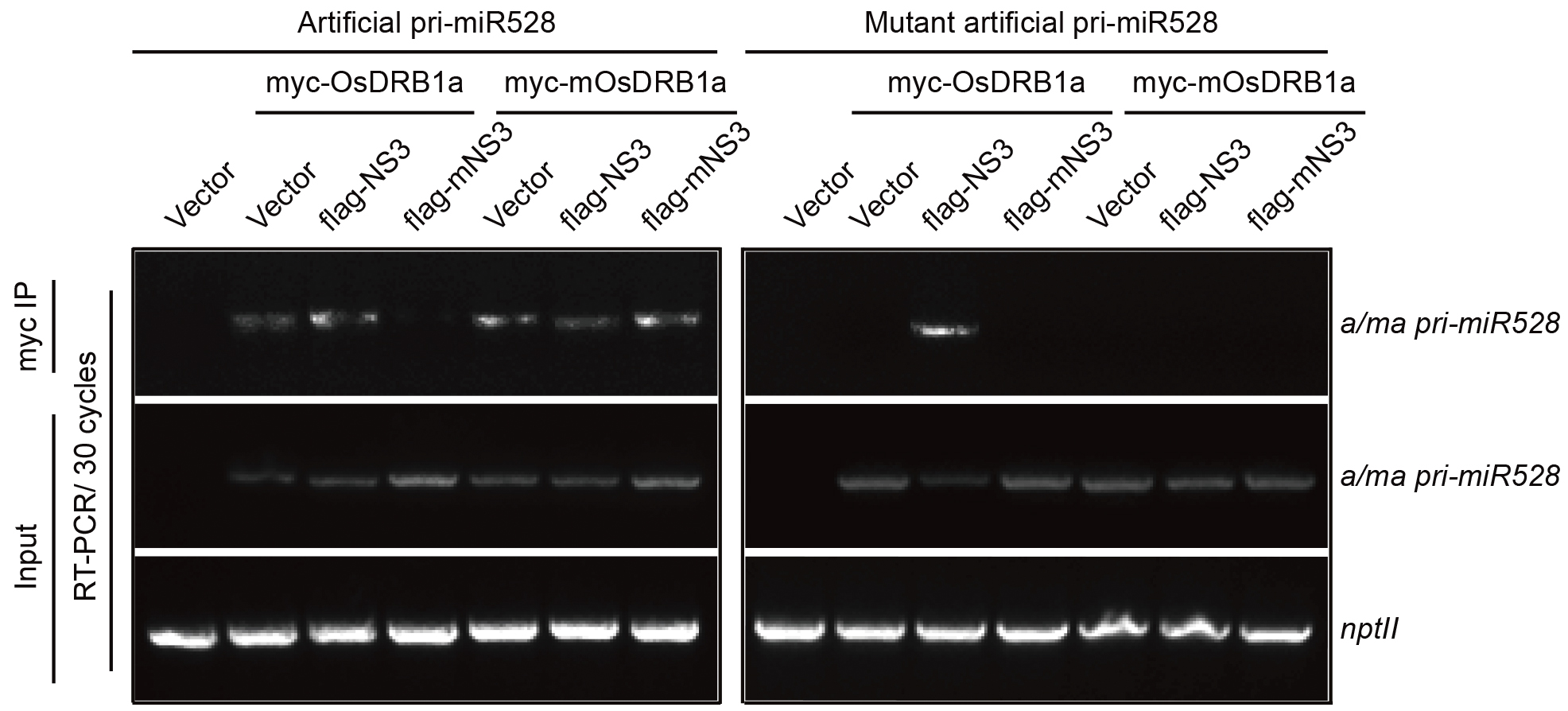
Figure 1. NS3 acts as a scaffold between DRB1 and pri-miRNA. RT-PCR detection of coimmunoprecipitated and input products of transiently co-expressed protein (DRB1a or mDRB1a), pri-miRNA (aprimiR528 or mapri-miR528), and protein (empty vector, NS3, or mNS3) in N. benthamiana. For more information, see Zheng et al., 2017.
Notes
- Keep all the buffers RNase-free.
- If you have many samples, isolates nuclei one by one, and freeze the nuclear pellet in the liquid nitrogen.
- More detailed information on nuclei isolation and immunoprecipitation can be obtained from Saleh et al., 2008 and Terzi et al., 2009.
Recipes
- Honda buffer
0.44 M sucrose
1.25% Ficoll
2.5% Dextran T40
20 mM HEPES-KOH, pH7.4
0.5% Triton X-100
10 mM MgCl2
20 U/ml RNase inhibitor
5 mM DTT (freshly added)
1x Cocktail Roche cOmplete (freshly added) - High salt nuclear lysis buffer
20 mM Tris-HCl, pH = 7.5
500 mM NaCl
4 mM MgCl2
20 U/ml RNase inhibitor (freshly added)
0.2% NP-40 (add fresh)
5 mM DTT (add fresh)
1x Cocktail Roche cOmplete (freshly added) - Dilution buffer
20 mM Tris-HCl, pH = 7.5
4 mM MgCl2
0.2% NP-40 (freshly added)
5 mM DTT (freshly added)
20 U/ml RNase inhibitor (freshly added)
1x Cocktail Roche cOmplete (freshly added) - IP buffer
20 mM Tris-HCl, pH = 7.5
100 mM NaCl
4 mM MgCl2
0.2% NP-40 (add fresh)
5 mM DTT (add fresh)
20 U/ml RNase inhibitor (add fresh)
1x Cocktail Roche cOmplete (freshly added)
Acknowledgments
This work was supported by grants to JG. W. and L. Y. from the National Natural Science Foundation of China (Nos. 31722045; 31772128; 31701757 and 31201491); the National Basic Research Program 973 (2014CB138400); and the Natural Science Foundation of Fujian Province of China, Outstanding Young Scientific Research Plan and Excellent Talent Plan in the New Century of Fujian Province (JA3091 and 2014J06011). This protocol was modified from Saleh et al., 2008 and Terzi et al., 2009. We declare no conflicts of interest or competing interests.
References
- Saleh, A., Alvarez-Venegas, R. and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3(6): 1018-1025.
- Terzi, L. C. and Simpson, G. G. (2009). Arabidopsis RNA immunoprecipitation. Plant J 59(1): 163-168.
- Zheng, L., Zhang, C., Shi, C., Yang, Z., Wang, Y., Zhou, T., Sun, F., Wang, H., Zhao, S., Qin, Q., Qiao, R., Ding, Z., Wei, C., Xie, L., Wu, J. and Li, Y. (2017). Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLoS Pathog 13(10): e1006662.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zheng, L., Zhang, C., Wu, J. and Li, Y. (2018). Detecting the Interaction of Double-stranded RNA Binding Protein, Viral Protein and Primary miRNA Transcript by Co-immunoprecipitation in planta. Bio-protocol 8(9): e2840. DOI: 10.21769/BioProtoc.2840.
Category
Microbiology > Microbe-host interactions > Virus
Plant Science > Plant molecular biology > Protein
Molecular Biology > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











