- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assessing the Efficacy of Small Molecule Inhibitors in a Mouse Model of Persistent Norovirus Infection
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2831 Views: 6557
Reviewed by: Yannick DebingKristin ShinglerBalaji Olety Amaranath

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Selection of Molecules with Immunological Potential from Excretory and Secretory Products from the Nematode Haemonchus placei by Cell Proliferation and Gene Expression Assays
Jocelyn Maza-Lopez [...] Carla O. Contreras-Ochoa
Jun 20, 2023 1320 Views

Detection of Cytoplasmic and Nuclear Circular RNA via RT-qPCR
Ke-En Tan [...] Yat-Yuen Lim
Sep 5, 2023 3391 Views

Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Ruben Warkentin and Anna Marie Pyle
Oct 20, 2025 1393 Views
Abstract
Human norovirus is the most common cause of acute gastroenteritis worldwide, resulting in estimated mortality of ~210,000 each year, of whom most are children under the age of five. However, norovirus can infect people of all age groups. There is a risk of prolonged infection in children, the elderly and patients who are immunocompromised. To study the inhibition of persistent norovirus replication by small molecule antivirals in vivo, we used a murine norovirus CR6 strain (MNV.CR6). We demonstrated earlier that efficient small molecules can reduce viral shedding in the stool of infected mice. Here we present how to generate the MNV.CR6 virus stock, infect type I and II interferon receptor knockout AG129 mice via oral gavage, administer antivirals to mice, and quantify viral genome copies in the stool of these mice.
Keywords: Murine norovirusBackground
Human noroviruses are an important cause of gastroenteritis. Although most norovirus infections are acute and self-limiting, the infection can become chronic in patients with an immunodeficient status, particularly in solid organ and hematopoietic stem cell transplant recipients, patients undergoing chemotherapy and patients with AIDS (Westhoff et al., 2009; Green, 2014; Angarone et al., 2016). Prolonged norovirus infection is also observed in young children and the elderly resulting in an increased duration of illness, increased defecations and virus shedding for up to 47 days (Murata et al., 2007; Aoki et al., 2010). Reduction of immunosuppressive therapy is, when feasible, the strategy of choice to control the infection in transplant recipients. Specific antiviral therapies to treat (chronic) norovirus gastroenteritis are not available. To assess the inhibitory effect of small molecules on persistent norovirus infections, we used a mouse-adapted persistent murine norovirus (MNV.CR6) in type I and II interferon receptor knockout AG129 mice (Strong et al., 2012). MNV is a genogroup V norovirus that is widely used as a surrogate for human noroviruses, which comprises around 30 strains (Karst et al., 2003; Wobus et al., 2004). The MNV.CR6 is avirulent in AG129 and STAT1-/- mice, but replicates for weeks to months in wild type mice and to higher titers in innate immune-deficient mice (Thackray et al., 2007). The MNV.CR6 strain has a tropism for the proximal colon and the cecum, where it persists and replicates more efficiently than the MNV.CW3 strain, which causes acute infection (Arias et al., 2012; Nice et al., 2013).
Materials and Reagents
- Overall
- Appropriate personal protection to work in a biosafety level 2 (BSL-2) laboratory or A-2 animal facility (gloves, lab coat, hairnet, shoe covers, safety glasses)
- Disinfectant: bleach (5,000 ppm) or Virkon S.
- Appropriate personal protection to work in a biosafety level 2 (BSL-2) laboratory or A-2 animal facility (gloves, lab coat, hairnet, shoe covers, safety glasses)
- In vitro work
- Culture flasks (150 cm2, TPP, catalog number: 90856 )
- Cell scrapers (Corning, Falcon®, catalog number: 353086 )
- Cryotubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 377224 )
- Pipet tips (10 µl, 100 µl, 1,000 µl)
- Disposable serological pipets (5 ml, 10 ml, 25 ml)
- Murine macrophage cells (RAW 264.7, ATCC, catalog number: TIB-71 )
- Dulbecco’s phosphate buffered saline (DPBS) (Thermo Fisher Scientific, catalog number: 14190094 )
- Dulbecco’s modified eagle’s medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 41965039 )
- Fetal calf serum (FCS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- Sodium bicarbonate (Thermo Fisher Scientific, GibcoTM, catalog number: 25080060 )
- L-Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030024 )
- HEPES (Thermo Fisher Scientific, GibcoTM, catalog number: 15630056 )
- Penicillin/streptomycin (P/S) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140148 )
- Sodium pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360039 )
- Culture medium (see Recipes)
- Culture flasks (150 cm2, TPP, catalog number: 90856 )
- In vivo work
- Eppendorf safe-lock tubes, 1.5 ml (Eppendorf, catalog number: 0030120086 )
- Syringe + needle for subcutaneous injection (VWR, catalog number: BDAM303176 )
- Needle container (Sharpsafe, catalog number: 41602432 )
- Bench surface protector (VWR, catalog number: 115-9220 )
- Earmarks (Bioseb, catalog number: EP-1005-1 )
- Plastic feeding tubes, 20 G x 30 mm, sterile (Instech Laboratories, catalog number: FTP-20-30 )
- Sterile 1 ml syringe that fits on the feeding tube (VWR, catalog number: 612-0106 )
- Plastic container to temporarily restrain a mouse
- Ear tags (Bioseb, catalog number: EP-1005-1 ) + applicator (Style 1005s1)
- Type I and II interferon receptor knockout AG129 mice (129/Sv mice), from BK Universal, UKCR6 strain
- 2’-C-methylcytidine (2CMC, Carbosynth, catalog number: NM07918 )
- Favipiravir (T-705, BOC Sciences, catalog number: B0084-463609 )
- Carboxymethylcellulose (CMC, Sigma–Aldrich, catalog number: C9481 )
- Sterile saline (NaCl 0.9%, B. Braun)
- Eppendorf safe-lock tubes, 1.5 ml (Eppendorf, catalog number: 0030120086 )
- Viral RNA quantification
- Multipette tip of 0.2 ml (Eppendorf, catalog number: 0030089413 )
- Lightcycler 480–Multiwell plate 96 (Roche Molecular Systems, catalog number: 04729692001 )
- RNeasy Mini kit (QIAGEN, catalog number: 74106 )
- Ethanol, absolute (Fisher Scientific, catalog number: 10342652 )
- iTaq Universal Probes One-Step Kit (Bio-Rad Laboratories, catalog number: 1725141 )
- Primers, probe and standard (Baert et al., 2008)
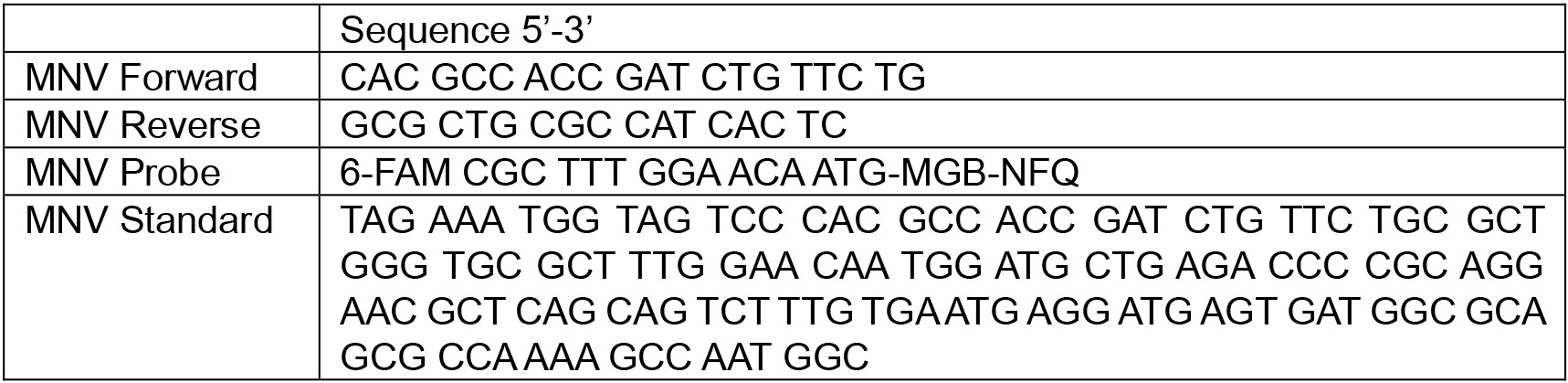
- Multipette tip of 0.2 ml (Eppendorf, catalog number: 0030089413 )
Equipment
- Biosafety hood in an A-2 animal facility
- Biosafety hood in a BSL-2 laboratory
- Incubator (37 °C, 5% CO2, humidified)
- Scale to weigh mice
- Forceps suitable for picking up mouse feces (autoclavable)
- Pipet set (P10, P100, P1000)
- Pipetboy (Integra Biosciences, catalog number: 155016 )
- Multipette® M4 (Eppendorf, catalog number: 4982000012 )
- Multichannel pipette (Eppendorf, catalog number: 3122000035 )
- -80 °C freezer
- PCR Workstation
- Vortex
- Centrifuge with a rotor suitable for 1.5 ml tubes
- Centrifuge with a rotor suitable for 50 ml tubes
- Autoclave
- RT-qPCR machine (Roche Diagnostics, model: Roche LightCycler® 96 )
- Inverted light microscope
- Cell counter
Software
- GraphPad Prism Software, version 7
- LightCycler® 96 SW 1.1 Software
Procedure
- Expansion and quantification of MNV.CR6 virus stock
Note: Culture and infect cells under a biosafety hood in a BSL-2 laboratory.- Split the RAW264.7 cells using a cell scraper, once a week at a 1:10 ratio in a T150 flask containing 40 ml of 10% media. Maintain the cells in a CO2 incubator at 5% CO2 and 37 °C.
- Infect the RAW264.7 cells with MNV.CR6 after 2 or 3 days of incubation i.e., when the cells are nearly confluent but not overgrown. Do this as follows:
- Aspirate the growth medium from cells, wash with DPBS and aspirate the DPBS.
- Add 1 ml of MNV.CR6 to the cells and distribute evenly, incubate for 1 h at 37 °C.
- Add 20 ml of 2% media and place the flask back in the incubator until the full cytopathic effect is observed (± 2-3 days post-infection).
- Aspirate the growth medium from cells, wash with DPBS and aspirate the DPBS.
- Repeat two freeze-thaw cycles (-80 °C vs. 37 °C), then collect the supernatants containing the virus after centrifugation (10 min, 1,500 x g) and store in aliquots (500-1,000 µl) in cryotubes at -80 °C.
- Determine the viral titer by endpoint titration as previously described in Reed et al. (1938). This procedure is based on the detection of cytopathic effect (CPE) by microscopy.
Note: Titration can be performed with a higher dilution series or extended over a second 96-well plate when the viral stock is strong.- Thaw MNV.CR6 stock at room temperature.
- Add 100 µl of 2% DMEM in all wells of a 96-well plate.
- Add 50 µl of pure virus stock in the wells B2-G2 of the second column of the 96-well plate.
- Homogenize the virus and medium with a multichannel pipette.
- Take 50 µl of the mixture and transfer into the next well.
- Repeat Steps A4d-A4e until reaching column 9 of the 96 well-plate.
- Homogenize the virus and medium in row 9 and discard 50 µl.
- Add 10,000 RAW 264.7 cells/well in a final volume of 100 µl in 2% DMEM, to the inner 60 wells.
- Incubate for 72 h in 5% CO2 at 37 °C.
- Compare the wells with infected cells (columns 2-9) to the wells with non-infected cells (columns 10-11) and evaluate for cytopathic effect (CPE) microscopically.
- Calculate TCID50 using the Reed-Muench formula (Reed et al., 1938).
- Thaw MNV.CR6 stock at room temperature.
- Split the RAW264.7 cells using a cell scraper, once a week at a 1:10 ratio in a T150 flask containing 40 ml of 10% media. Maintain the cells in a CO2 incubator at 5% CO2 and 37 °C.
- Oral gavage of MNV.CR6 in mice
Notes:- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as close as possible to a specific pathogen free (SPF) facility.
- Use a protective bench coat to prevent viral contamination of the biosafety hood.
- Replace gloves between different experimental groups.
- For all experiments the mice were age- and sex-matched, mice were 8-12 weeks of age.
- Tag an ear of each mouse with a unique number.
- Thaw virus and dilute in 2% media to 106 CCID50 (50% cell culture infectious dose) of MNV.CR6. Give each mouse 200 μl of virus via oral gavage (see below).
- Fill the syringe and feeding tube with virus and remove all air bubbles.
- Carefully pick up the mouse by the base of its tail, place onto a wire cage.
- With the other hand, restrain the mouse by holding the scruff between thumb and forefinger. Place the tail between the little finger and ring finger to stretch the mouse (Figure 1).

Figure 1. Correct hand position to hold a mouse for oral gavage - Now gently insert the feeding tube vertically down into the esophagus and administer virus in a steady motion (Figure 2). Any resistance felt indicates improper placement of the feeding tube, in which case take out the feeding tube and re-position.

Figure 2. Vertical insertion of the feeding tube in the esophagus of the mouse
- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as close as possible to a specific pathogen free (SPF) facility.
- Administration of small molecule inhibitors
Notes:- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as closely as possible to an SPF facility.
- Use a protective bench coat to prevent viral contamination of the biosafety hood.
- Replace gloves between different experimental groups.
Subcutaneous injection of 2’-C-Methylcytidine (100 mg/kg/day)- Dissolve 2CMC in sterile saline.
- Fill the syringe with virus and remove all air bubbles.
- Hold the mouse by the base of its tail and place it onto a wire cage.
- Insert the needle parallel to the skin on the back of the mouse. Make sure you are just under the skin (Figure 3).

Figure 3. Subcutaneous injection in the back of the mouse - Inject the mouse with 2CMC via the subcutaneous route in a steady motion with a volume (~200 µl) that has a final concentration of 100 mg/kg/day.
Oral gavage of favipiravir (T-705) (200 mg/kg/day)
- Dissolve T-705 in 0.4% CMC.
- Administer the appropriate volume of T-705 via oral gavage with a volume (~200 µl) that has a final concentration of 200 mg/kg/day.
- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as closely as possible to an SPF facility.
- Follow up and stool collection
Notes:- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as close as possible to an SPF facility.
- Use a protective bench coat to prevent viral contamination of the biosafety hood.
- Decontaminate boxes, forceps and scale between different experimental groups.
- Evaluate the general condition of the animal on a daily basis and check the overall health (weight, behavior, fur). If an animal reaches the defined humane endpoints (loses more than 15% of body weight in 1-2 days or an overall of > 20% in body weight or displays obvious signs of suffering [lethargy, squinted eyes, dehydration, hunched back]), it has to be humanely euthanized according to the European guidelines (Dir. 2010/63/EU).
- Place each mouse individually in a clean plastic container awaiting the defecation.
- Use a forceps to collect one piece of feces and place it in a labeled 1.5 ml tube (Figure 4). Repeat for all mice.

Figure 4. Collection of a stool sample in a labeled tube - Score the feces samples for consistency according to the table below.

- Store stool samples in a -20 °C or -80 °C freezer.
- Handle infected animals under a biosafety hood in an A-2 animal facility under conditions as close as possible to an SPF facility.
- Viral RNA extraction from stool samples
Note: Handle infectious samples under a biosafety hood in a BSL-2 laboratory.- Extract the viral RNA using the RNeasy mini kit of QIAGEN:
- Add 400 µl of RLT buffer to each feces sample.
- Vortex the samples very well until the feces sample is completely homogenized.
- Spin the tubes for 3 min at 10,000 x g.
- Transfer 350 µl of the supernatants to a new 1.5 ml tube and add 350 µl of 70% ethanol.
- Continue according to manufacturer’s protocol.
- Add 400 µl of RLT buffer to each feces sample.
- Elute the viral RNA in 50 µl RNase free water.
- Store the viral RNA in a -20 °C freezer.
- Extract the viral RNA using the RNeasy mini kit of QIAGEN:
- Quantification of viral RNA by RT-qPCR
Note: Prepare the mixtures for the RT-qPCR in a PCR workstation.- Prepare the correct concentrations of primers, probe and the standard dilution (108 to 100 RNA copies).
- Thaw viral RNA samples and components of the one-step iTaq Universal Probes kit.
- Prepare for each sample a one-step RT-qPCR mix, a total of 16 µl containing: 10 µl iTaq Universal Probes reaction mix, 0.5 µl of iScript advanced reverse transcriptase, RNase free water, 900 nM of MNV primers and 200 nM of MNV probe.
- Gently vortex and briefly centrifuge the mix.
- In each well of the 96 RT-qPCR well plate, pipet 16 µl of the mix (using a multipipette) and add 4 µl of viral RNA.
- Close the RT-qPCR plate properly with a plastic seal.
- Place the RT-qPCR plate in a LightCycler and run with the following cycling conditions: reverse transcription at 50 °C for 10 min, initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 15 sec, annealing and extension at 60 °C for 30 sec.
- For the absolute quantification, use a 10-fold dilution series of the MNV DNA standard with known concentration.c
- Prepare the correct concentrations of primers, probe and the standard dilution (108 to 100 RNA copies).
Data analysis
Viral RNA copy numbers of RNA extracts are determined using the standard curve and the LightCycler® 96 SW 1.1 Software. The amount of viral RNA copy numbers per gram of feces can be calculated by weighing the stool samples before extraction. Statistical analysis was performed using Prism 7 software, P-values were determined with the nonparametric Mann-Whitney test. For representative details see our original publication (Rocha-Pereira et al., 2015).
Notes
- AG129 mice (129/Sv mice) were originally obtained from BK Universal, UK, and under an annual license agreement bred and housed at our institute under SPF conditions.
- All experiments using mice strictly followed the guidelines of and were performed with the approval of the Ethical Committee of the KU Leuven, Belgium (P101/2012).
Recipes
- Culture media for RAW264.7 cells

Acknowledgments
The study in which the protocol described here, has been published previously (Rocha-Pereira et al., 2015). The development and use of this protocol were supported by the EU FP7 project SILVER (260644), KU Leuven IOF project HB/14/031 and GOA grant (GOA/10/014). J.Rocha-Pereira and the research leading to these results has received fundingfrom the People Programme (Marie Curie Actions) of the European Union's SeventhFramework Programme (FP7/2007-2013) under REA grant agreement no 608765. J. Van Dycke is an SB Ph.D. fellow of the Scientific Fund for Research of Flanders (FWO). We gratefully acknowledge Prof. Herbert W. Virgin (Washington University, St. Louis, USA) for kindly providing the MNV.CR6 virus. We thank Carolien De Keyzer, Jasper Rymenants and Charlotte Vanderheydt for excellent technical assistance. The authors declare no conflicts of interest or competing interests.
References
- Angarone, M. P., Sheahan, A. and Kamboj, M. (2016). Norovirus in transplantation. Curr Infect Dis Rep 18(6): 17.
- Aoki, Y., Suto, A., Mizuta, K., Ahiko, T., Osaka, K. and Matsuzaki, Y. (2010). Duration of norovirus excretion and the longitudinal course of viral load in norovirus-infected elderly patients. J Hosp Infect 75(1): 42-46.
- Arias, A., Bailey, D., Chaudhry, Y. and Goodfellow, I. (2012). Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. J Gen Virol 93(Pt 7): 1432-1441.
- Baert, L., Wobus, C. E., Van Coillie, E., Thackray, L. B., Debevere, J. and Uyttendaele, M. (2008). Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl Environ Microbiol 74(2): 543-546.
- Green, K. Y. (2014). Norovirus infection in immunocompromised hosts. Clin Microbiol Infect 20(8): 717-723.
- Karst, S. M., Wobus, C. E., Lay, M., Davidson, J. and Virgin, H. W. t. (2003). STAT1-dependent innate immunity to a Norwalk-like virus. Science 299(5612): 1575-1578.
- Murata, T., Katsushima, N., Mizuta, K., Muraki, Y., Hongo, S. and Matsuzaki, Y. (2007). Prolonged norovirus shedding in infants <or=6 months of age with gastroenteritis. Pediatr Infect Dis J 26(1): 46-49.
- Nice, T. J., Strong, D. W., McCune, B. T., Pohl, C. S. and Virgin, H. W. (2013). A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J Virol 87(1): 327-334.
- Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27(3): 493-7.
- Rocha-Pereira, J., Van Dycke, J. and Neyts, J. (2015). Treatment with a nucleoside polymerase inhibitor reduces shedding of murine norovirus in stool to undetectable levels without emergence of drug-resistant variants. Antimicrob Agents Chemother 60(3): 1907-1911.
- Strong, D. W., Thackray, L. B., Smith, T. J. and Virgin, H. W. (2012). Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J Virol 86(6): 2950-2958.
- Thackray, L. B., Wobus, C. E., Chachu, K. A., Liu, B., Alegre, E. R., Henderson, K. S., Kelley, S. T. and Virgin, H. W. t. (2007). Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol 81(19): 10460-10473.
- Westhoff, T. H., Vergoulidou, M., Loddenkemper, C., Schwartz, S., Hofmann, J., Schneider, T., Zidek, W. and van der Giet, M. (2009). Chronic norovirus infection in renal transplant recipients. Nephrol Dial Transplant 24(3): 1051-1053.
- Wobus, C. E., Karst, S. M., Thackray, L. B., Chang, K. O., Sosnovtsev, S. V., Belliot, G., Krug, A., Mackenzie, J. M., Green, K. Y. and Virgin, H. W. (2004). Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2(12): e432.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dycke, J. V., Neyts, J. and Rocha-Pereira, J. (2018). Assessing the Efficacy of Small Molecule Inhibitors in a Mouse Model of Persistent Norovirus Infection. Bio-protocol 8(9): e2831. DOI: 10.21769/BioProtoc.2831.
Category
Microbiology > in vivo model > Viruses
Molecular Biology > RNA > qRT-PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









