- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Glycogen and Extracellular Glucose Estimation from Cyanobacteria Synechocystis sp. PCC 6803
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2826 Views: 7080
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simple Methods for the Preparation of Colloidal Chitin, Cell Free Supernatant and Estimation of Laminarinase
Ananthamurthy Koteshwara
Oct 5, 2021 5152 Views

Extraction and Electrophoretic Analysis of Bacterial Lipopolysaccharides and Outer Membrane Proteins
Yue-Jia Lee and Thomas J. Inzana
Dec 20, 2021 5751 Views

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3552 Views
Abstract
Cyanobacteria, which have the extraordinary ability to grow using sunlight and carbon dioxide, are emerging as a green host to produce value-added products. Exploitation of this highly promising host to make products may depend on the ability to modulate the glucose metabolic pathway; it is the key metabolic pathway that generates intermediates that feed many industrially important pathways. Thus, before cyanobacteria can be considered as a leading source to produce value-added products, we must understand the interaction between glucose metabolism and other important cellular activities such as photosynthesis and chlorophyll metabolism. Here we describe reproducible and reliable methods for measuring extracellular glucose and glycogen levels from cyanobacteria.
Keywords: Extracellular glucoseBackground
Cyanobacteria have a light-dark cycle in their natural habitat. In the light, their metabolism is centered on photosynthesis, the Calvin cycle, glycolysis and the TCA cycle with N-assimilation; carbon is stored as glycogen. In the dark, glycogen is metabolized through glycolysis and the oxidative pentose phosphate (OPP) pathway, the oxidative and reductive branches of the TCA cycle, and the C4 cycle (Nagarajan et al., 2014). Thus, the shift from dark to light or light to dark drives metabolic reprogramming.
In the laboratory, the addition of glucose to the culture media also impacts cyanobacteria metabolic programs. For example, nutritional and environmental conditions influence how the cyanobacterium Synechocystis metabolizes glucose; Synechocystis metabolizes glucose differently in photoautotrophic, heterotrophic and mixotrophic conditions. Previous studies reported that some strains of Synechocystis are light-dependent and glucose tolerant (Anderson and McIntosh, 1991). Light-activated heterotrophic growth (LAHG) conditions are characterized by the presence of glucose and growth in the dark with a pulse of white or blue light for at least 5-15 min per day. However, some strains of Synechocystis are glucose intolerant, meaning that they cannot grow in the presence of glucose in the dark. In summary, the addition of glucose to the culture media of Synechocystis has been reported to bring physiological and metabolic changes such as pigmentation (Ryu et al., 2004), carbon metabolism (Lee et al., 2007; Takahashi et al., 2008), phosphorylation patterns (Bloye et al., 1992), carbon dioxide uptake (Kaplan and Reinhold, 1999), and oxidative stress generation (Narainsamy et al., 2013).
To identify the utility of cyanobacteria to produce natural product, growing cyanobacteria in large-scale is a prerequisite. For growing cyanobacteria efficiently, it’s important to characterize the direct impact of common environmental factors such as light and temperature on glucose metabolism. Here, we present an accurate, reproducible, and reliable method to quantify extracellular glucose and glycogen levels of cyanobacteria, we belelive that this method will help determine the utility of cyanobacteria as a source for engineering natural products.
Materials and Reagents
- Pipette tips (20 µl-1 ml, autoclaved)
- Aluminum foil
- 1.5 and 2 ml Eppendorf tubes (autoclaved)
- 0.45 µm filter
- Cyanobacteria Synechocystis sp. PCC 6803 (WT, mutant D95 & C95)
Note: For more information about these strains, please see Data analysis A4. - Sulfuric acid (ACS reagent, Sigma-Aldrich, catalog number: S1526 )
- Nitrogen
- Ethanol (Sigma-Aldrich, catalog number: 362808-1L )
- Glycogen (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0561 )
- Amyloglucosidase (Sigma-Aldrich, catalog number: 10115-1G-F )
- Sodium carbonate (Na2CO3) (Sangon Biotech, catalog number: ST0840 )
- Sodium nitrate (NaNO3) (Sinopharm Chemical Reagent, catalog number: 10019918 )
- Hydrochloric acid (HCl) (Sinopharm Chemical Reagent, catalog number: 10011018 )
- Sodium hydroxide (NaOH) (Sangon Biotech, catalog number: SB0617 )
- Potassium phosphate dibasic (K2HPO4) (Sangon Biotech, catalog number: PB0447 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sangon Biotech, catalog number: MT0864 )
- Ferric ammonium sulphate (Sangon Biotech, catalog number: A502657 )
- Citric acid (Sangon Biotech, catalog number: C0529 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sangon Biotech, catalog number: CT1331 )
- EDTA-Na2 (Sangon Biotech, catalog number: E0105 )
- Boric acid (H3BO3) (Sangon Biotech, catalog number: BB0044 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Sangon Biotech, catalog number: A500331 )
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sangon Biotech, catalog number: A602906 )
- Sodium molybdate dehydrate (Na2MoO4·2H2O) (Sangon Biotech, catalog number: SB0865 )
- Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sangon Biotech, catalog number: A501425 )
- Cobalt(II) nitrate hexahydrate (Co(NO3)2·6H2O) (Sangon Biotech, catalog number: CB7774 )
- Sodium thiosulfate anhydrous (Na2S2O3) (Sangon Biotech, catalog number: S1712 )
- D-glucose (Sangon Biotech, catalog number: 501991 )
- Glucose standard solution (Sigma-Aldrich, catalog number: G3285 )
- Benzoic acid (Sinopharm Chemical Reagent, catalog number: 30018615 )
- Glucose oxidase/peroxidase (Sigma-Aldrich, catalog number: G3660 )
- o-Dianisidine reagent (Sigma-Aldrich, catalog number: D2679 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: P6310 )
- Sodium acetate (Sigma-Aldrich, catalog number: S2889 )
- TES, N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (Sangon Biotech, catalog number: TB0927 )
- BG-11 media (see Recipes)
- D-Glucose (see Recipes)
- Assay reagent (see Recipes)
- 30% (w/v) KOH (see Recipes)
- 100 mM sodium acetate (pH 4.5) (see Recipes)
Equipment
- Conical flasks (100, 200, 500 ml) (SHUNIU, Chengdu, China)
- Vortex (FINE PCR, model: Finevortex )
- Centrifuge machine, unrefrigerated, maximum speed 17,000 x g, with rotor for microtubes (Thermo Fisher Scientific, model: HeraeuesTM PicoTM 17 )
- Glass test tubes 18 x 150 mm
- Spectrophotometer (METASH, model: V-5600 )
- 1 ml glass cuvettes (METASH)
- Pipettes (20 µl, 200 µl, 1 ml, 5 ml) (Gilson, France)
- Water baths (temperature set at 37 ± 1 °C; 60 °C; 92 °C) (Meier, model: XMTD-204 )
- Shaking light-incubator (light:up to 150 µmoles/m2/sec; temperature: 20-50 °C) (Shanghai Zhichu, model: ZQWY-200G )
- Freeze dryer (Labconco, model: FreeZone Plus 6 )
- Oven, 60 °C (Boxun, model: GZX-9140MBE )
- Autoclave (Boxun, model: YXQ-LS-100SII )
- pH meter (Mettler Toledo, model: FE 20 )
- Swimming holders
Software
- GraphPad PRISM (Version 5.01)
Procedure
- Cultivation of cyanobacteria
- Cyanobacterium (Synechocystis) cell stocks are kept at -80 °C in 20% glycerol. Add ~100 µl of the frozen stock to 50 ml BG-11 media (see Recipe 1) in a 100 ml conical flask. Grow a starter culture for 3-4 days (until OD730 is around 0.5) under white light (25 μmol m-2 sec-1) in a light-incubator at 30 °C with shaking at 150 rmp. To conduct glucose consumption and glycogen estimation experiments, make a fresh culture in 100 ml BG-11 media in a 200 ml conical flask with an initial OD730 of 0.1 and incubate in a photo-incubator under the same conditions mentioned above.
- When the OD730 is around 0.4 (it usually takes ~36 h), 1 ml (0.5 M) sterile D-glucose solution (see Recipe 2) is added to the 100 ml culture media to get a final glucose concentration of 5 mM. Grow cultures in two conditions: (1) Dark-glucose condition, in which the conical flask is wrapped with aluminum foil and (2) Light-glucose condition in which the conical flask is not wrapped in foil. Collect a sample for glucose and glycogen measurement every 24 h from both conditions.
- For each 2 ml collection, transfer it to a 2 ml Eppendorf tube, measure the OD730 using a spectrophotometer and record it. Centrifuge the 2 ml samples at 17,000 x g for 2 min at room temperature and transfer the supernatant to a separate 2 ml Eppendorf tube. The supernatant is used for assaying glucose and the rest is used for assaying glycogen (more details in glycogen measurement section).
- Cyanobacterium (Synechocystis) cell stocks are kept at -80 °C in 20% glycerol. Add ~100 µl of the frozen stock to 50 ml BG-11 media (see Recipe 1) in a 100 ml conical flask. Grow a starter culture for 3-4 days (until OD730 is around 0.5) under white light (25 μmol m-2 sec-1) in a light-incubator at 30 °C with shaking at 150 rmp. To conduct glucose consumption and glycogen estimation experiments, make a fresh culture in 100 ml BG-11 media in a 200 ml conical flask with an initial OD730 of 0.1 and incubate in a photo-incubator under the same conditions mentioned above.
- Extracellular glucose measurement
- The sample from Step A3 is used to assay for glucose (Table 1)
Table 1. Experimental design for extracellular glucose test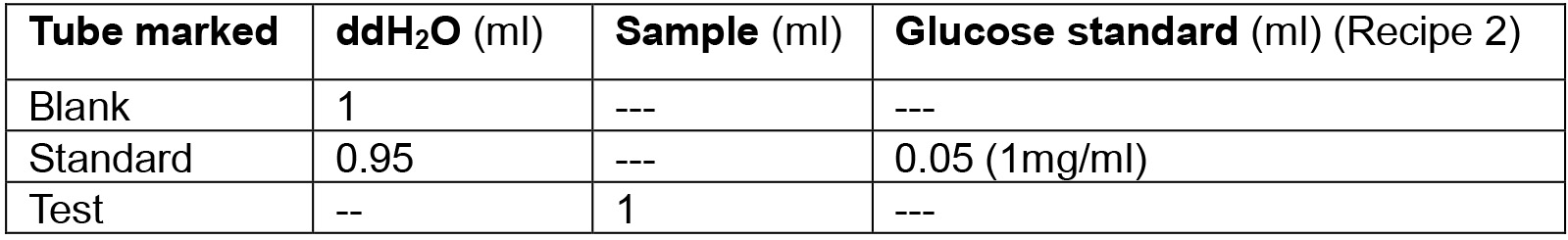
Note: We take 5 biological & 3 technical replicates for each experiment. - Open the spectrophotometer. Add 2 ml assay reagent (see Recipe 3) to each tube. Mix thoroughly by vortexing. Add 500 µl of samples and standard to cuvettes and measure the absorbance at 540 nm against the blank. Record as ‘initial reading’.
- Place the tubes in a water bath at 37 °C for exactly 30 min. Stop the reaction by adding 2 ml of 12 N H2SO4 (add the acid in the chemical hood for safety) and mix thoroughly with extra care by vortexing. To get the exact incubation duration, keep a 30-60 sec interval of pipetting of blank, standard and test samples. The presence of glucose results in the development of pink color (Figures 1A and 1B). The more glucose in the sample, the stronger the pink color looks.
- The OD at 540 is measured after 30 min against the blank and recorded as ‘final reading’.
Note: The Step B4 should be performed immediately after Step B3. The time interval should be maintained strictly equal.
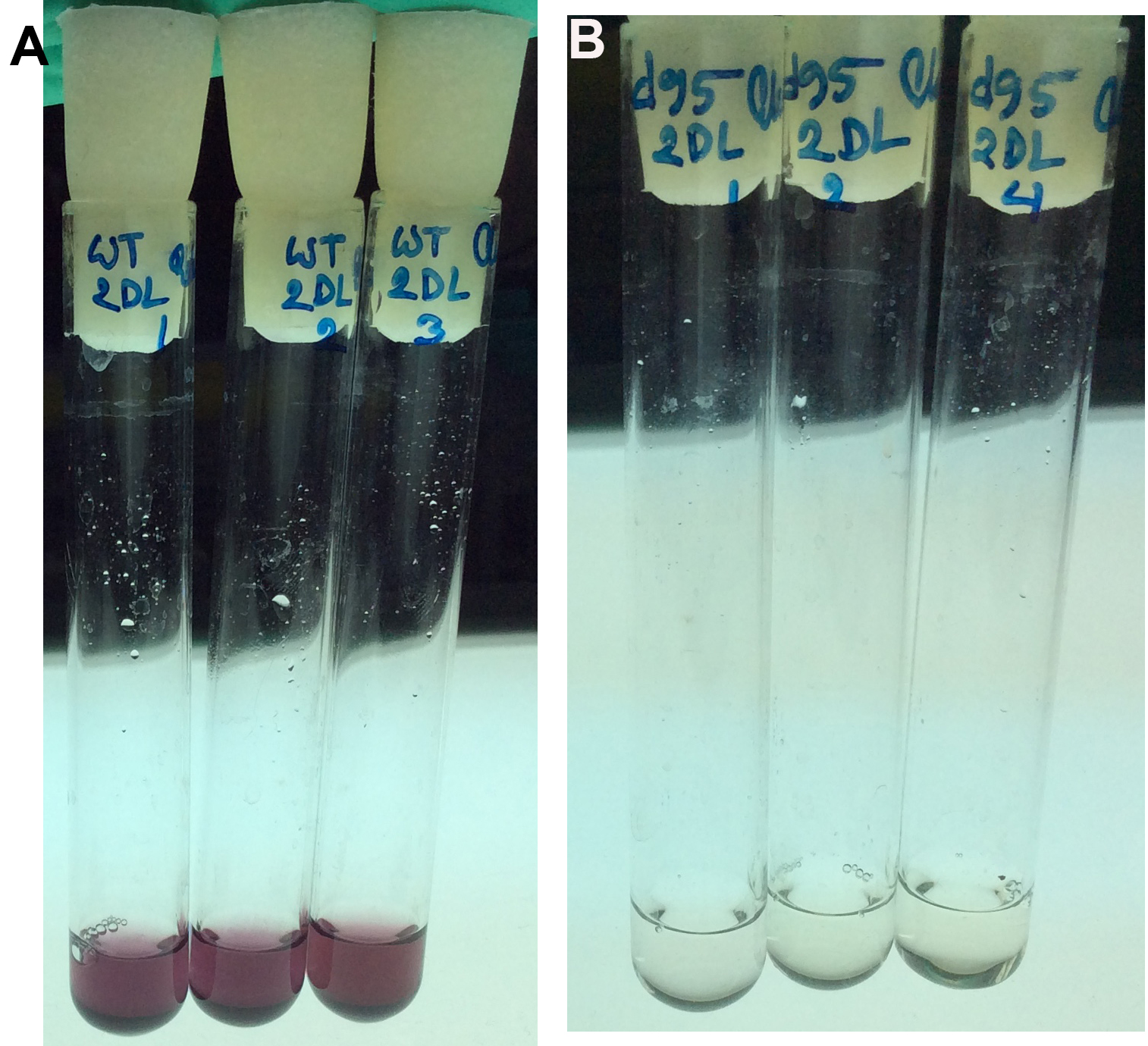
Figure 1. Quantitative assay of extracellular glucose from growth medium. Three replicates are shown for WT (panel A) and a mutant cyanobacterium (panel B). In principle, glucose is oxidized to gluconic acid and hydrogen peroxide by glucose oxidase. Hydrogen peroxide then reacts with reduced o-Dianisidine in the presence of peroxidase to form a brown colored oxidized o-Dianisidine. Oxidized o-Dianisidine reacts with sulfuric acid to form a more stable pink-colored product. The intensity of the pink color measured at 540 nm is proportional to the original glucose concentration. Thus, the pink color indicates the presence of glucose, whereas the white/clear color indicates very little glucose. The results are also available in Khan et al., 2016. - The sample from Step A3 is used to assay for glucose (Table 1)
- Glycogen measurement
- Use the sample collected in Step A3. Calculate the approximate cell number from the OD730 value as follows; OD730 value of 1.0 can be taken as 1.03 x 108 cells per ml. Centrifuge approximately 3 x 109 cells from wild type and mutant Synechocystis cultures each at 1,700 x g for 2 min. Remove the supernatant. Wash the pellet each with 1 ml sterile ddH2O two times. Immediately, move the cell pellet to -80 °C for 5 min to stop the metabolism.
- Next, place the cell pellet in a freeze dryer at -70 °C overnight under a continuous flow of nitrogen. On the next day, after drying ~14 h in the freeze dryer, weigh the dried cell pellet and record the weight.
- Resuspend the dried cell pellet in 1 ml of 30% (w/v) KOH (see Recipe 4). Mix completely by pipetting (3 to 5 min).
- Place the mixture in a 97 °C water bath for 2 h with swimming holder (set the water bath in advance to make sure the temperature is correct).
- Divide the 1 ml of 30% KOH-suspended sample into two separate Eppendorf tubes, each with 500 µl. Add ~1.3 ml ice cold ethanol to a final concentration of 70-75%. Incubate on ice for 2 h.
- Centrifuge the mixture at 17,000 x g for 2 min at room temperature. Remove the supernatant while being careful not to disturb the pellet at the bottom of the tube. Wash the pellet three times with 98% ethanol. The pellet seen at the bottom of the Eppendorf tube is glycogen.
- Open the lid of the Eppendorf tube and place in a 60 °C oven for 10-20 min (depends on how long it takes to dry completely).
- Resuspend the dried pellet in 0.5 ml 100 mM sodium acetate (pH 4.5) (see Recipe 5). If two tubes contain the same sample (divided in Step C5), they can be added together to get a volume of 1 ml. Dissolve a fixed amount of glycogen (0.005 mg) in 1 ml 100 mM sodium acetate (pH 4.5). This is the STDglyc.
- Add 2 mg amyloglucosidase to the 1 ml suspension obtained in the Step C8 (final concentration 2 mg/ml). Incubate in a 60 °C water bath for 2 h. Amyloglucosidase hydrolyzes glycogen into glucose.
- Measure the glucose following Steps B4 to B7 described in ‘Procedure B’.
- Use the sample collected in Step A3. Calculate the approximate cell number from the OD730 value as follows; OD730 value of 1.0 can be taken as 1.03 x 108 cells per ml. Centrifuge approximately 3 x 109 cells from wild type and mutant Synechocystis cultures each at 1,700 x g for 2 min. Remove the supernatant. Wash the pellet each with 1 ml sterile ddH2O two times. Immediately, move the cell pellet to -80 °C for 5 min to stop the metabolism.
Data analysis
- Extracellular glucose measurement
- The difference between the ‘final reading’ and ‘initial reading’ is calculated for the standard (ΔA540STD) and test samples (ΔA540Sample). The amount of glucose can be calculated using the equation below:
Mg glucose = (ΔA540Sample) x (amount of glucose in 0.05 ml standard solution/ΔA540STD). - The amount of glucose obtained is converted into mMol and normalized by dividing the OD730 recorded in Step A3. For each sample, we use five biological and three technical replicates.
- Open the GraphPad PRISM (Version 5.01) software (the interface of the software is shown in Figure 2). Select ‘grouped’ from the ‘New Graph & Table’ list. Select ‘start with an empty data table’ from the ‘Sample data’ option. Select ‘Interleaved Bar, Vertical’ from the “Choose a Graph’ option. From the ‘Y subcolumns for replicates or error bars’ option, select ‘Enter’ with 5 replicates. Select ‘Mean with SEM’ from the ‘Plot’ option and then click on ‘create’.

Figure 2. The interface of the GraphPad PRISM software - A page will open with 5 empty boxes on the top under the single title A (A:Y1, A:Y2, A:Y3, A:Y4, A:Y5). A series of titles will appear on the left marked with 1, 2, 3, 4, etc. Replace the label of title “A” with “WT”, “B” with “D95” and “C” with “C95”. The left title series are labeled with the corresponding time points: 0 h, 24 h, etc. The mean value of the three technical replicates is put in the box and there are five boxes for five biological replicates. Likewise, data for D95 and C95 are also placed in the boxes marked with title D95 and C95. D95 is a mutant in which the slr089 gene has been deleted, which results in this mutant having different responses to glucose metabolism compared to wild type. C95 is a strain where the endogenous slr0895 gene is deleted, but it is complemented with a wild type version of the slr089 gene linked to a different antibiotic resistance marker that was used to delete the slr089 gene in strain D95. C95 has wild type glucose metabolic activity. Analyses are performed by clicking the ‘Analysis’ option on the top of the page. The statistics can be viewed by clicking the ‘Result’ option on the left. The graph can be viewed by clicking on the ‘Graph’ option on the left of the page. Figure 3A can be copied by right clicking and selecting copy.
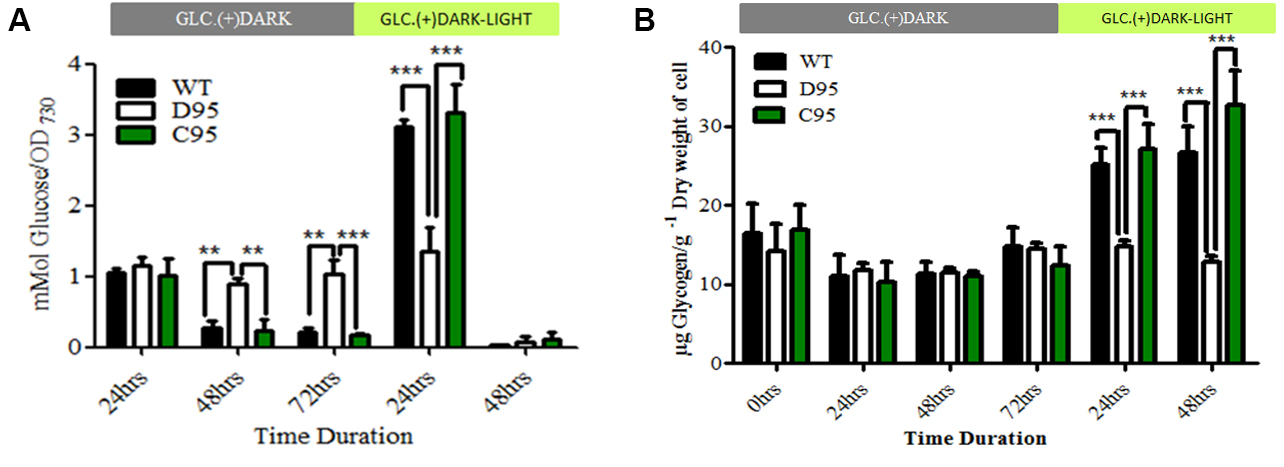
Figure 3. Data analysis and presentation of extracellular glucose (A), and glycogen (B) estimation from wild type and different mutant strains of cyanobacteria, Synechocystis sp. PCC 6803 (see details in Data analysis A4). The results have been published in Scientific Report and more information on the mutants can be found in Khan et al. (2016).
Schematic presentation of the reaction: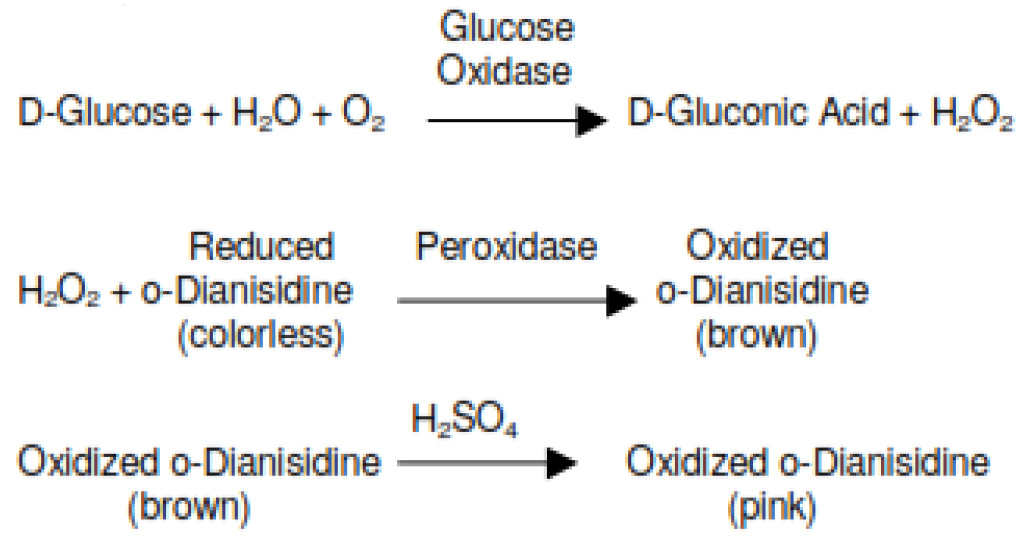
- The difference between the ‘final reading’ and ‘initial reading’ is calculated for the standard (ΔA540STD) and test samples (ΔA540Sample). The amount of glucose can be calculated using the equation below:
- Glycogen measurement from cyanobacteria
- The amount of glycogen is calculated using the formula below:
Amount of glycogen (mg) in test sample = (amount of glucose of test sample) x (0.005/amount of glucose obtained from STDglyc).
The amount of glycogen calculated is multiplied by 1,000 to convert the unit from mg to µg. - The glycogen calculated in µg is normalized by dividing by the dry weight measured in Step C2 (Procedure section). To get reliable data, we use five biological and three technical replicates for each sample including the glycogen standard (STDglyc). We make an average of glucose obtained from STDglyc replicates before using in Step C1.
- Statistical analyses are performed following Steps A3 to A5 in the Data analysis section and sample results are shown in Figure 3B.
- The amount of glycogen is calculated using the formula below:
Recipes
- BG-11 media preparation
- Step 1: The stock solutions are prepared according to the composition given in Table 2 which were based on the solutions reported by Stainer et al. (1971). Autoclave the stock solutions. Wrap stock 3 with aluminum foil to protect from light. Store the stock solutions at room temperature
Table 2. BG-11 media composition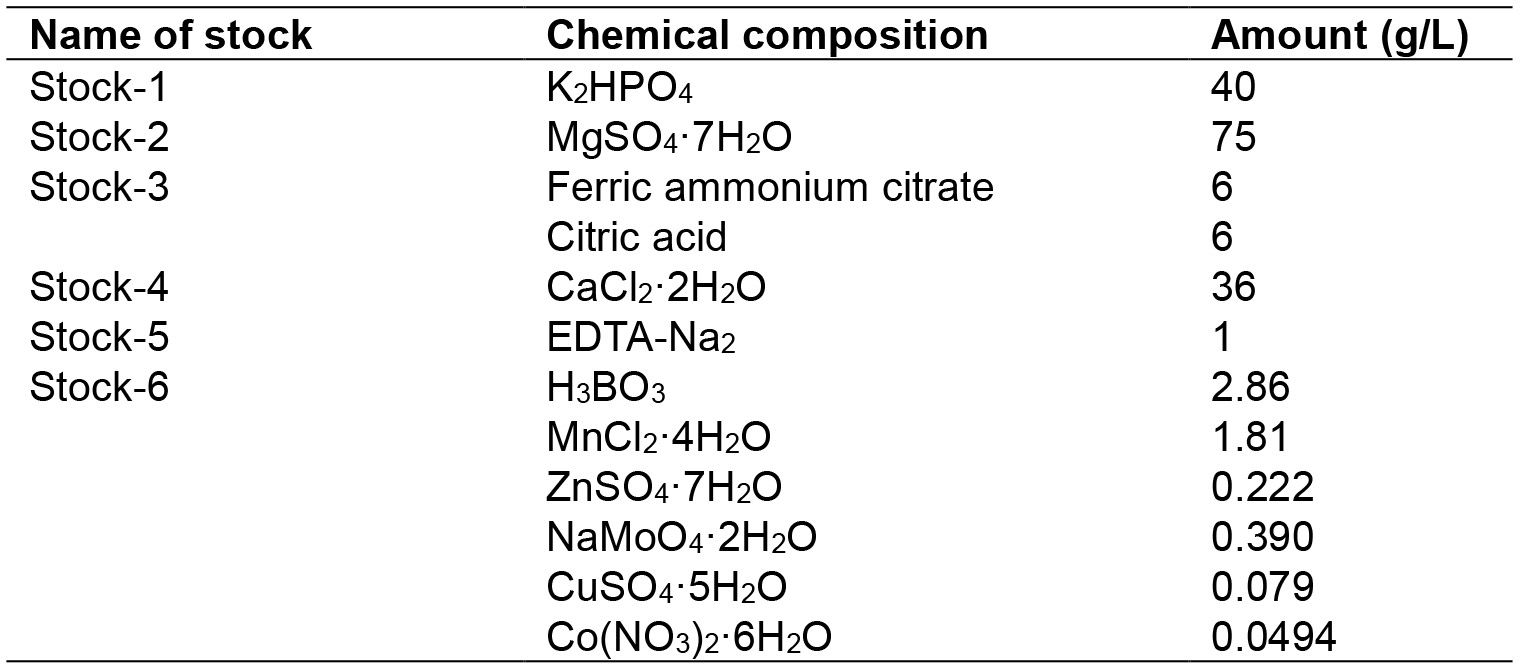
- Step 2: To prepare 1 L of media, add 1 ml from each stock. Next, add 0.02 g/L of Na2CO3 and 1.5 g/L of NaNO3. The pH was adjusted to ~7.5 using hydrochloric acid (HCl) and sodium hydroxide (NaOH)
- Step 3: Pour the media into a conical flask and autoclave using the liquid media cycle
- Step 1: The stock solutions are prepared according to the composition given in Table 2 which were based on the solutions reported by Stainer et al. (1971). Autoclave the stock solutions. Wrap stock 3 with aluminum foil to protect from light. Store the stock solutions at room temperature
- D-Glucose
Dissolve 1.350 g D-glucose in 15 ml of dH2O to make 0.5 M stock
Sterilize using a 0.45 µm filter - Assay reagent
Add 0.8 ml of the o-Dianisidine reagent to an amber bottle containing 39.2 ml of glucose oxidase/peroxidase reagent
Invert the bottle several times to mix
Minimize exposure to light
This solution is stable for up to 1 month at 2-8 °C
Discard if turbidity develops or color forms - 30% (w/v) KOH preparation
Dissolve 15 g of KOH in 50 ml ddH2O to prepare a 30% (w/v) KOH solution
Store at room temperature - 100 mM sodium acetate preparation
Dissolve 0.4102 g sodium acetate in 50 ml water to get a final concentration of 100 mM
Adjust the pH to 4.5 using NaOH or HCl
Store at room temperature
Acknowledgments
This protocol is an adapted version of the method described by Grundel et al. (2012). This work was supported by the 973 Program (2011CBA00803), the National Natural Science Foundation of China (31671504, 81421061), and the National Key Technology R&D Program (2012BAI01B09).
Authors declare no any conflicts of interest or competing interests.
References
- Anderson, S. L. and McIntosh, L. (1991). Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J Bacteriol 173(9): 2761-2767.
- Bloye, S. A., Silman, N. J., Mann, N. H. and Carr, N. G. (1992). Bicarbonate concentration by Synechocystis PCC6803: Modulation of protein phosphorylation and inorganic carbon transport by glucose. Plant Physiol 99(2): 601-606.
- Grundel, M., Scheunemann, R., Lockau, W. and Zilliges, Y. (2012). Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158(Pt 12): 3032-3043.
- Kaplan, A. and Reinhold, L. (1999). CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50: 539-570.
- Khan, R. I., Wang, Y. S., Afrin, S., Wang, B., Liu, Y., Zhang, X. Q., Chen, L., Zhang, W. W., He, L. and Ma, G. (2016). Transcriptional regulator PrqR plays a negative role in glucose metabolism and oxidative stress acclimation in Synechocystis sp. PCC 6803. Sci Rep 6: 32507.
- Lee, S., Ryu, J. Y., Kim, S. Y., Jeon, J. H., Song, J. Y., Cho, H. T., Choi, S. B., Choi, D., de Marsac, N. T. and Park, Y. I. (2007). Transcriptional regulation of the respiratory genes in the cyanobacterium Synechocystis sp. PCC 6803 during the early response to glucose feeding. Plant Physiol 145(3): 1018-1030.
- Nagarajan, S., Srivastava, S. and Sherman, L. A. (2014). Essential role of the plasmid hik31 operon in regulating central metabolism in the dark in Synechocystis sp. PCC 6803. Mol Microbiol 91(1): 79-97.
- Narainsamy, K., Cassier-Chauvat, C., Junot, C. and Chauvat, F. (2013). High performance analysis of the cyanobacterial metabolism via liquid chromatography coupled to a LTQ-Orbitrap mass spectrometer: evidence that glucose reprograms the whole carbon metabolism and triggers oxidative stress. Metabolomics 9(1): 21-32.
- Ryu, J. Y., Song, J. Y., Lee, J. M., Jeong, S. W., Chow, W. S., Choi, S. B., Pogson, B. J. and Park, Y. I. (2004). Glucose-induced expression of carotenoid biosynthesis genes in the dark is mediated by cytosolic pH in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 279(24): 25320-25325.
- Takahashi, H., Uchimiya, H. and Hihara, Y. (2008). Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry. J Exp Bot 59(11): 3009-3018.
- Stanier, R. Y., Kunisawa, R., Mandel, M. and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35(2): 171-205.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Khan, M. R. I., Wang, Y., Afrin, S., He, L. and Ma, G. (2018). Glycogen and Extracellular Glucose Estimation from Cyanobacteria Synechocystis sp. PCC 6803. Bio-protocol 8(9): e2826. DOI: 10.21769/BioProtoc.2826.
Category
Microbiology > Microbial biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Glycogen
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









