- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Thrips Damage Using Ilastik and ImageJ Fiji
Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2806 Views: 10838
Reviewed by: Pengpeng LiEugenio LlorensOliver Xiaoou Dong

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Maize Seedlings Colonization with Serendipita indica and Its Colonization Efficiency Analysis
Om Prakash Narayan [...] Atul Kumar Johri
Oct 20, 2023 2267 Views

Workflow for a Functional Assay of Candidate Effectors From Phytopathogens Using a TMV-GFP-based System
Peng Cao [...] Yuyan An
Apr 20, 2025 1712 Views
Abstract
Quantification of insect damage is an essential measurement for identifying resistance in plants. In screening for host plant resistance against thrips, the total damaged leaf area is used as a criterion to determine resistance levels. Here we present an objective novel method for analyzing thrips damage on leaf disc using the freely available software programs Ilastik and ImageJ. The protocol was developed in order to screen over 40 Capsicum lines for resistance against Frankliniella occidentalis (Western Flower Thrips) and Thrips tabaci (Onion thrips).
Keywords: Insect resistanceBackground
Quantification of insect damage is an essential measurement for identifying resistance in plants. In screening programs for host-plant resistance against thrips, the total damaged leaf area is used as a criterion to determine resistance levels. Thrips damage is characterized by silvery spots that show high contrast with the intact leaf area, but the feeding spots also include darker areas ranging from dark green to brown. These gradual discolorations of the leaf are too subtle to precisely quantify with programs such as Winfolia (http://www.regentinstruments.com/assets/winfolia_software.html) or ImageJ (Rasband, 2011) alone. As a result, thrips damage is commonly scored by individuals that rate the samples. Samples are classified into categories signifying the amount of damage (Mirnezhad et al., 2010; Maharijaya et al., 2011 and 2012), or damage is estimated to the nearest 1 mm2 (Leiss et al., 2009; Mirnezhad et al., 2010; Maharijaya et al., 2011 and 2012). These subjective measurements make comparison between studies/screening programs difficult. Moreover, they are time consuming and thus costly for breeding companies. Here we present an objective high-throughput standardized screening method to measure leaf surface damage caused by thrips using the freely available software programs ImageJ Fiji (Schindelin et al., 2012) and Ilastik (Sommer et al., 2011). Ilastik has a wide range of applications ranging from cell biology (Fabrowski et al., 2013), where it is used to compute the amount of surface flattening of epithelial cells, to biomechanics (Bongiorno et al., 2014), where it is used to identify boundaries of human mesenchymal stem cells. It is an easy-to-use, self-learning image processing program that allows segmentation and classification of two-dimensional surfaces based on labels provided by the user (Sommer et al., 2011). ImageJ is often used to quantify the amount of removed leaf area by chewing herbivores and the total leaf surface of intact leaves (Meyer and Hull-Sanders, 2008; Morrison and Lindell, 2012). However, it is rarely used to quantify feeding damage caused by thrips. Thrips feeding causes rather subtle discolorations on the leaves. ImageJ is limited in quantifying such color differences, for which Ilastik provides a more suitable alternative.
Materials and Reagents
- Filtration paper (e.g., filtration paper nr 600, VWR, catalog number: 516-0309 )
- Ziploc® like bags (e.g., 18 x 25 cm, 50 µm polyethylene foliezak met druksluiting, Vink Lisse, catalog number: 174718 49 )
- Petri Dish diameter 15 cm (e.g., non-treated culture dishes, Corning, catalog number: 430597 )
- Parafilm® M (e.g., BRAND®, Parafilm®, VWR, catalog number: 291-1213 )
- Glass jar (Figure 1) (e.g., 555 ml Twist-off pot TO82 with Twist-off deksel RTS82 wit BPA NI lid, www.glazenflessenenpotten.nl, GLAZEN RLESSEN EN POTTEN. NL, catalog numbers: V2391WS and VC305 )

Figure 1. Glass jar for thrips starvation (height 11.5 cm x diameter 7.5 cm) - Synchronized L1/L2 Frankliniella occidentalis (Pergande) or Thrips tabaci (Lindeman) (Thysanoptera)
- Capsicum annuum (Solanaceae) plants (any variety)
- Agar (e.g., Phyto Agar, Duchefa Biochemie, catalog number: P1003 )
- Water
- 1.5% liquid agar solution (see Recipes)
Equipment
- Climate cabinet set to 25 °C for F. occidentalis or 23 °C for T. tabaci, L16:D8 light regime (e.g., Economic Delux 432 L with TL lightning, Snijders Labs, http://www.snijderslabs.com)
- Microwave (Moulinex, model: Micro-chef FM2515Q )
- Cork borer, diameter 1.5 cm (e.g., Humboldt Brass Cork Borer Set with Handles, Fisher Scientific, catalog number: 07-865-10B)
Manufacturer: Humboldt Mfg., catalog number: H-9663 . - Beaker 50 ml (e.g., Griffin beakers, Corning, PYREX®, catalog number: 1000-50 )
- Soft paint brush (e.g., van Eyck paint brush set, brush #1)
- Plastic tweezers (e.g., Azlon Forceps - Tweezers, Dynalab Corp., Dynalon Labware, catalog number: 516555-0001 , https://www.dynalon.com/PublicStore/)
- Epson 10000XL scanner (Epson, model: 10000XL ) or any SLR camera (12 mega pixel) with tripod
- Handmade grid with 2 cm spacing (Figure 3)
- Black paper (for scanner) or black cloth (for SLR camera)
- Laptop with installed software
- Precision balance (Sigma-Aldrich, catalog number: Z267074)
Manufacturer: Sartorius, model: BP 310 S . - Laboratory bottle with cap 500 ml (DWK Life Sciences, Duran, catalog number: 21 801 44 5 )
Software
- ImageJ Fiji e.g., version 2.0.0 with Java 1.6.0_24
- Ilastik version 1.1.3,
Note: For successful application of the protocol, it is important to use this exact version. The software can be found online: files.Ilastik.org/1.1/, ‘ilastik-1.1.3-win64.exe’, also available for Linux and OSX. - Epson Scan Utility e.g., version 3.4.9.9
Note: Only necessary if an Epson scanner is used or equivalent when using another scanner.
Procedure
- Thrips preparation
Collect synchronized L1/L2 larvae in a glass jar lined with 3 pieces of slightly moist filtration paper 24 h prior to the experiments. Place the glass jar in a climate cabinet set at 25 °C for F. occidentalis or 23 °C for T. tabaci, with a L16:D8 light regime. This 24 h period of starvation ensures that thrips will start feeding directly after the no-choice leaf disc assay is started. - No-choice leaf disc assay
- Prepare 1.5 % liquid agar solution (see Recipe 1)
- Collect leaves from the plants in the greenhouse and transport to the laboratory in closed Ziploc-like plastic bags containing 2 ml of water to keep the leaves fresh.
- Heat the agar in a microwave until it is liquefied.
- Use a cork borer to punch leaf discs while avoiding the mid-vein. Midveins often have light color patches that might result in inaccurate thrips damage quantification.
- Pour part of the agar solution in a beaker and pour a small drop (approximate 2 cm Ø) of liquid agar in the middle of a Petri dish (Figure 2A). Place the leaf disc on the agar with the adaxial side in full contact with the agar. This ensures that the thrips can only feed on the abaxial side (‘underside’) of the leaf. Place the leaf disc on the agar just before the agar becomes solid (gelling temperature between 34-36 °C, Duchefa Biochemie Safety Data Sheet P1003 version 2.0).
- Place five thrips on each leaf disc with a small paintbrush (Figure 2B). Make sure that the agar is fully solid before placing the thrips on the leaf disc.
- Close the Petri dishes with Parafilm® M and place them in a climate cabinet for 48 h (Figure 2C).
- After 48 h, the thrips can be removed with a small paintbrush and digital images of the leaf discs can be acquired.

Figure 2. Experimental setup of leaf disc assay. Placing the leaf disc on the agar (A), inoculating with thrips (B) and placing the closed Petri dishes in a tray in the climate cabinet (C).
- Prepare 1.5 % liquid agar solution (see Recipe 1)
- Acquiring the images
- Place the leaf discs with plastic tweezers on a scanner (or on black cloth in the case an SLR camera is used).
- For easy processing of the acquired image, all leaf discs should be equally distributed, so that leaf discs can be easily ‘cut out’ using a single macro run in ImageJ Fiji. Equal distribution of the leaf discs can be achieved using a grid (Figure 3).
- The total number of objects (leaf discs and labels) placed on the horizontal and vertical axis should be equal (in our case: horizontal axis: 3 leaf disc + 1 label, vertical axis: 4 leaf discs).
- When you use the protocol, it is advisable to include leaf discs that have not been exposed to thrips damage (control leaf discs) and analyze these in the same way as the leaf discs exposed to thrips. Ilastik sometimes overestimates the amount of damage, for example, due to the presence of leaf veins. The undamaged leaf disc images can be used to correct for these errors.
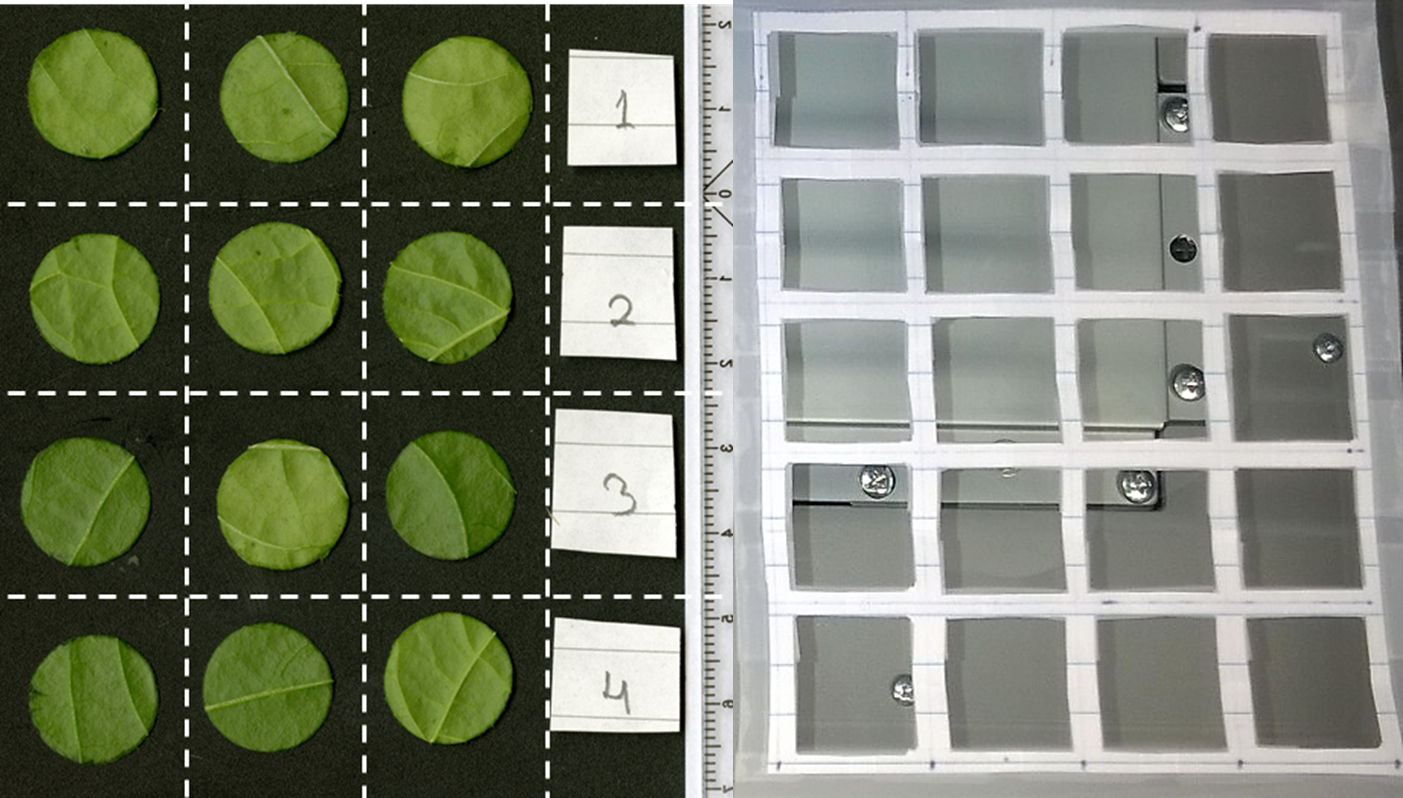
Figure 3. Placement of leaf discs. A. Example of TIFF image using a scanner set to 1,200 dpi. Horizontal and vertical axis are marked for illustration with white dashed lines in this example. A ruler is included at the right side of the image for calibration. B. Example of a handmade paper grid that can be used to equally distribute single leaf discs on a scanner.
- For easy processing of the acquired image, all leaf discs should be equally distributed, so that leaf discs can be easily ‘cut out’ using a single macro run in ImageJ Fiji. Equal distribution of the leaf discs can be achieved using a grid (Figure 3).
- Cover the leaf discs placed on the scanner with black paper and obtain a TIFF image of the leaf discs with the scanner set to 1,200 dpi. Include a calibration square (1 x 1 cm) or a ruler in one of the scans that can be used for calibration (x pixels = x mm). When an SLR camera is used, take pictures from a given distance (with a calibration square (1 x 1 cm or a ruler)) in JPEG or JPG format.
- Place the leaf discs with plastic tweezers on a scanner (or on black cloth in the case an SLR camera is used).
- Cutting scan image in ImageJ Fiji
- Open the scan image acquired in Step C2 in ImageJ Fiji. Click ‘File’ and select ‘Open...’ or drag your selected images from one folder directly to ImageJ Fiji.
- Open a macro, click on ‘plugins’; ‘new’; ‘macro’ (Figure 4).
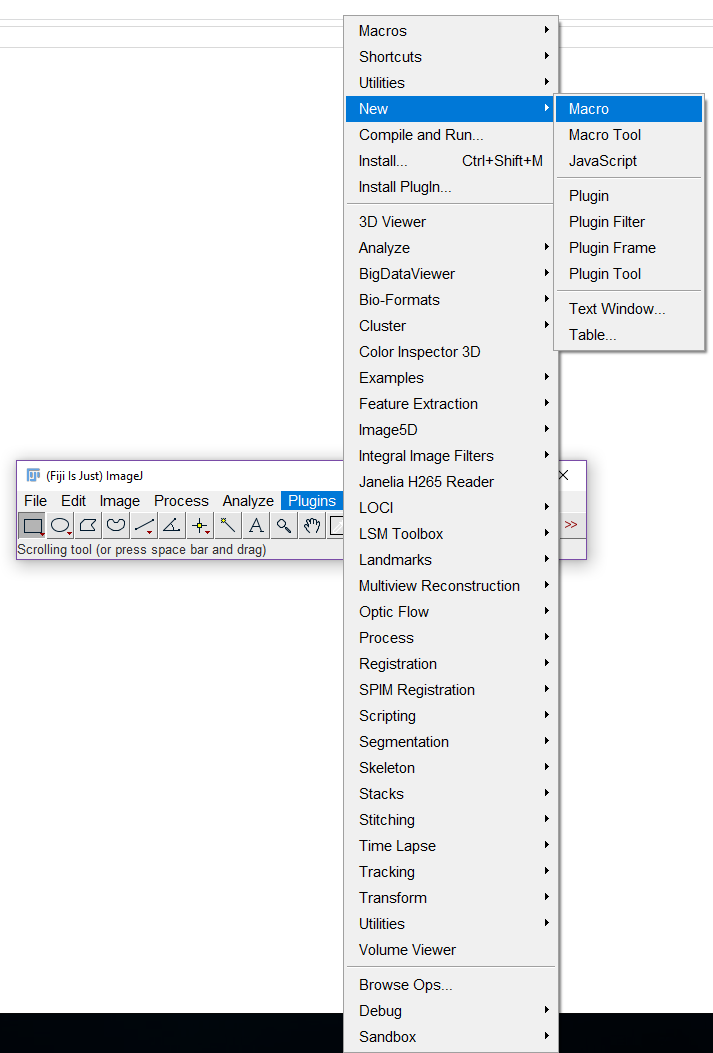
Figure 4. Opening a new macro in ImageJ Fiji - Copy the following script (only black text) in the macro.ijm.ijm window (Figure 5). Text highlighted in green are comments from the authors and should not be included in the macro. Text in red should be completed by the user.
n = getNumber(X,X);
//fill in the number of cuts that has to be made on the spots of the red ‘X’. In case of our acquired scan image: n = getNumber(4,4).
id = getImageID();
title = getTitle();
getLocationAndSize(locX, locY, sizeW, sizeH);
width = getWidth();
height = getHeight();
tileWidth = width / n;
tileHeight = height / n;
for (y = 0; y < n; y++) {
offsetY = y * height / n;
for (x = 0; x < n; x++) {
offsetX = x * width / n;
selectImage(id);
call("ij.gui.ImageWindow.setNextLocation", locX + offsetX, locY + offsetY);
tileTitle = title + " [" + x + "," + y + "]";
run("Duplicate...", "title=" + tileTitle);
makeRectangle(offsetX, offsetY, tileWidth, tileHeight);
run("Crop");
}
}
//The images have been cut, but each obtained slice should have a unique name
imageCount = nImages
for (image = 1; image <= imageCount; image++) {
selectImage(image);
// Changes the title of the active image to a string name
rename("test" + image);
}
//Saves all the slices in the directory you want (marked in red below), make sure to use two backslashes between the folder names.
n=nImages;
for(i=0,1; i<n; i++){
title=getTitle;
saveAs("TIFF", "X:\\...\\.....\\" + title);
close();
}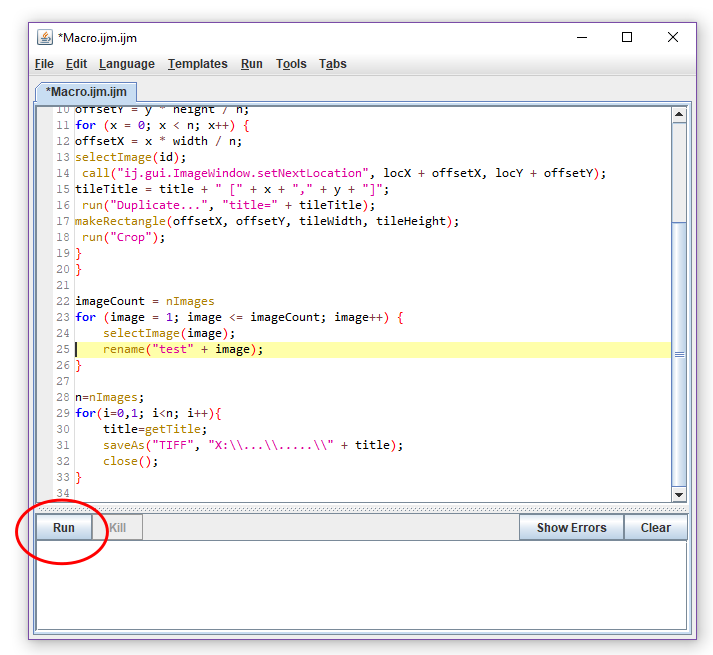
Figure 5. Running the macro. The macro cuts the scan image in ImageJ Fiji to obtain images (slices) with single leaf discs. - Run the macro by clicking ‘Run’. ImageJ Fiji now automatically cuts out and saves the single leaf discs as TIFF files in the directory that you have chosen. The single leaf discs are labeled with the title of the scan and a number (Titelscanimage_1, Titelscanimage_2, etc.). ImageJ Fiji starts to cut out leaf discs from the top left corner of the first row, continues to the right and then starts with the next row.
- Images produced from the macro (Figure 6) can be further analyzed in Ilastik

Figure 6. Single leaf disc after cutting scan image with ImageJ Fiji
- Open the scan image acquired in Step C2 in ImageJ Fiji. Click ‘File’ and select ‘Open...’ or drag your selected images from one folder directly to ImageJ Fiji.
- Training Ilastik to recognize thrips damage
- Open Ilastik and create a new project by selecting ‘Pixel classification’ and give your project a name.
- Now import the scans with the single leaf discs by clicking ‘Add New’ and select ‘Add separate image(s)…’ (Figure 7). You can now select the images on your computer that should be imported in Ilastik. Image import takes 3.26MB/sec with an Intel® coreTM i7-4910MQ CPU @ 2.90 GHz, RAM 16 GB (as a reference 50 images of each 3.08 MB take 48 sec to be imported).
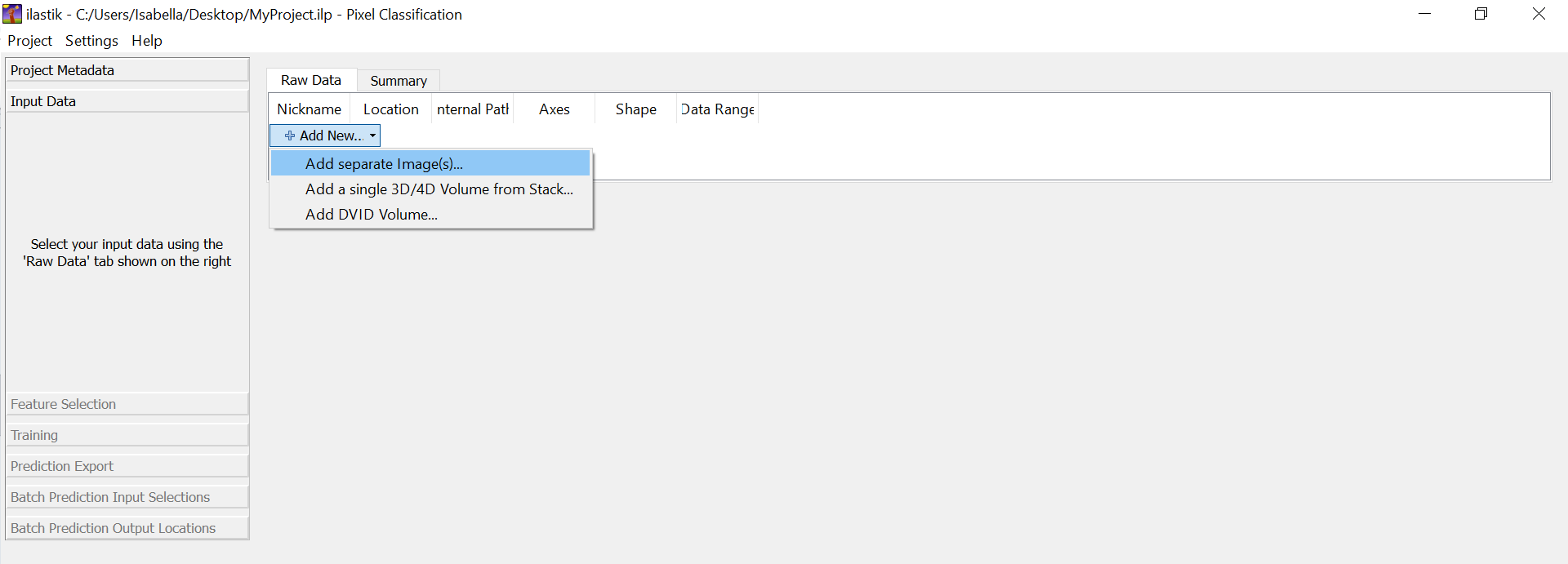
Figure 7. Importing images to Ilastik - You can now select features to use for your analysis. Click on the tab ‘Select Features’ and select features. We recommend selecting color/intensity/texture on ơ = 1.0 px (Figure 8).
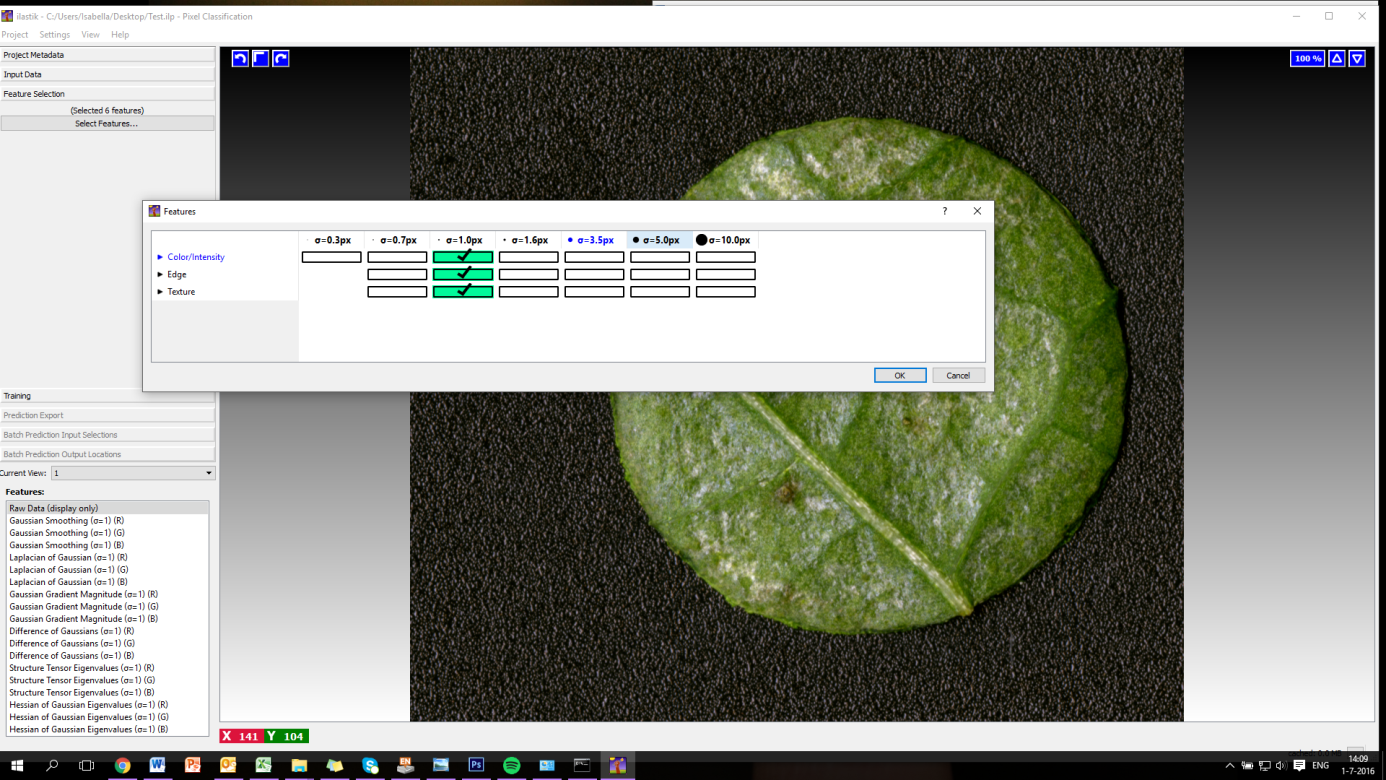
Figure 8. Setting parameters for image analysis in Ilastik - Go to the tab ‘Training’, select ‘Add label’ and add 3 labels. One will be for the background, one for the leaf disc and one for the actual thrips damage. The colors will be assigned automatically by the program. Name your labels as shown in Figure 9. Make sure that you keep the same order as shown in the screenshot (Figure 9); this is of importance for later analysis.
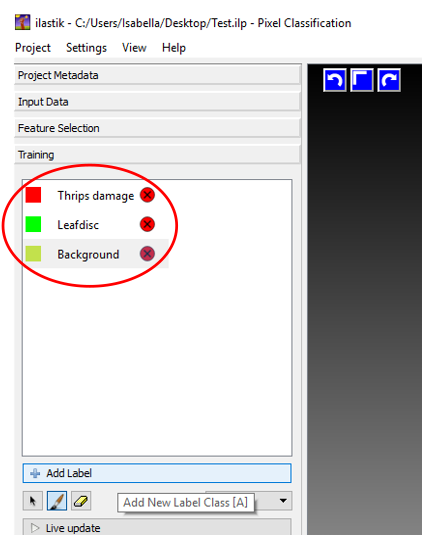
Figure 9. Adding segmentation labels - Now you can start with the training of the program. Select one of the labels and mark the areas that correspond with the labels. It is important to use red to mark areas that are typical thrips damage, with green undamaged leaf areas and with yellow the dark background (Figure 10A). Make sure that you use several leaf discs for the training of the program. You can switch to other leaf discs by choosing the picture number or name from the drop down list next to ‘Current view:’ (Figure 10B). Training is an import step, since the program depends on sufficient training to accurately recognize the different components in the image. Training can take up to half an hour, with a training image processing speed of approximately 0.9 MB/sec (Intel® coreTM i7-4910MQ CPU @ 2.90 GHz, RAM 16 GB). As a reference, approximate 10 cm (with the pencil tool set to 3 pixels) of thrips damaged area marking is necessary for accurate learning.
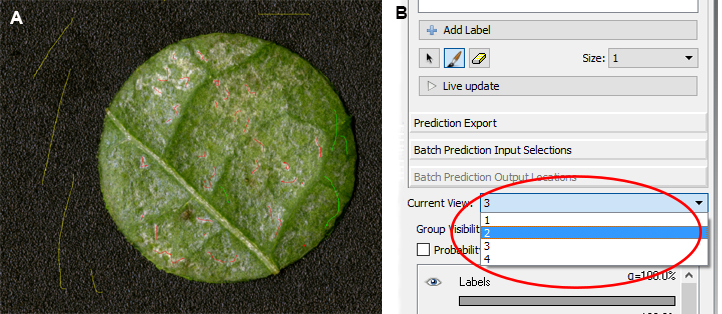
Figure 10. Training phase in Ilastik. A. Marking of thrips damage (red), the leaf disc (green) and the background (yellow); B. Switching between imported images in Ilastik. - To check whether the training has been sufficient, click on ‘Live update’. The areas are now colored corresponding to what the program sees as thrips damage (red/pink), the leaf disc (green) and the background (yellow) (Figure 11). If the program does not sufficiently distinguish between thrips damage, leaf disc and background, click again on ‘Life update’ and uncheck ‘Probability’ (marked with redcircle, Figure 11). Add more markers in the picture as specified above and activate ‘Life update’ again. It is advisable to train the program separately for dark and light green leaves, e.g., different leaf ages, accessions or species, to ensure optimal results. In Figure 12 an example of sufficient and insufficient training of Ilastik is provided.
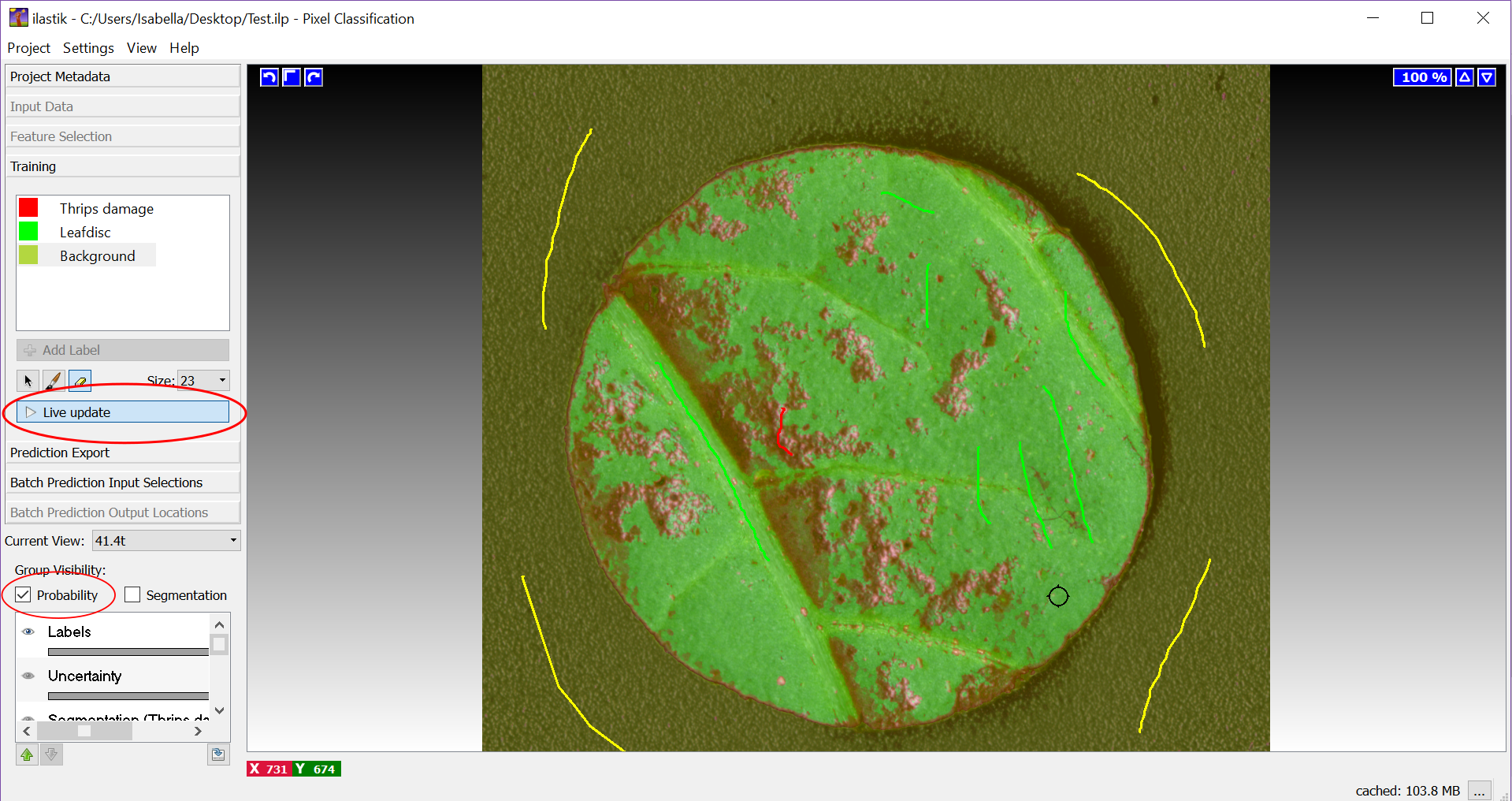
Figure 11. Live view of image segmentation in the training stage in Ilastik (A)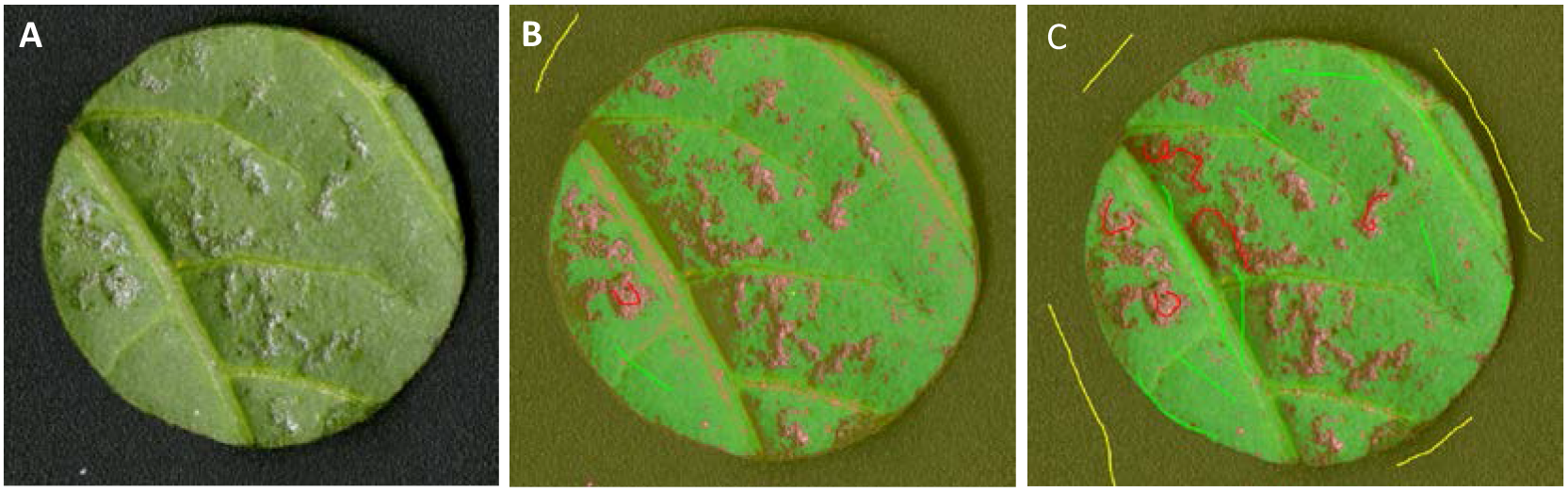
Figure 12. Illustration of training in Ilastik. A. The original leaf disc; B. After insufficient training of the leaf disc. In B, thrips damage is overestimated e.g., leaf veins are incorrectly marked as thrips damage and the area right from the leaf vein is also seen as incorrect thrips damage. C. Sufficient training of the leaf disc after marking more area with the correct labels in Ilastik.
- Open Ilastik and create a new project by selecting ‘Pixel classification’ and give your project a name.
- Converting images into simple segmentations
After the training is complete, all imported images can be converted to JPEG files that are simple segmentations (black, grey and white image) of the original pictures.- Go to the tab ‘Prediction Export’, select by ‘Export sources’ ‘Simple segmentation’.
- Now go the ‘Export Settings’ and click on ‘Choose settings’. Make sure the settings are the same as in Figure 13. Datatype should be set to ‘signed 8-bit’ and the output file format ‘JPEG’. Make sure you select the location that you want to save your output files in. After you make sure the settings are correct, click ‘OK’.
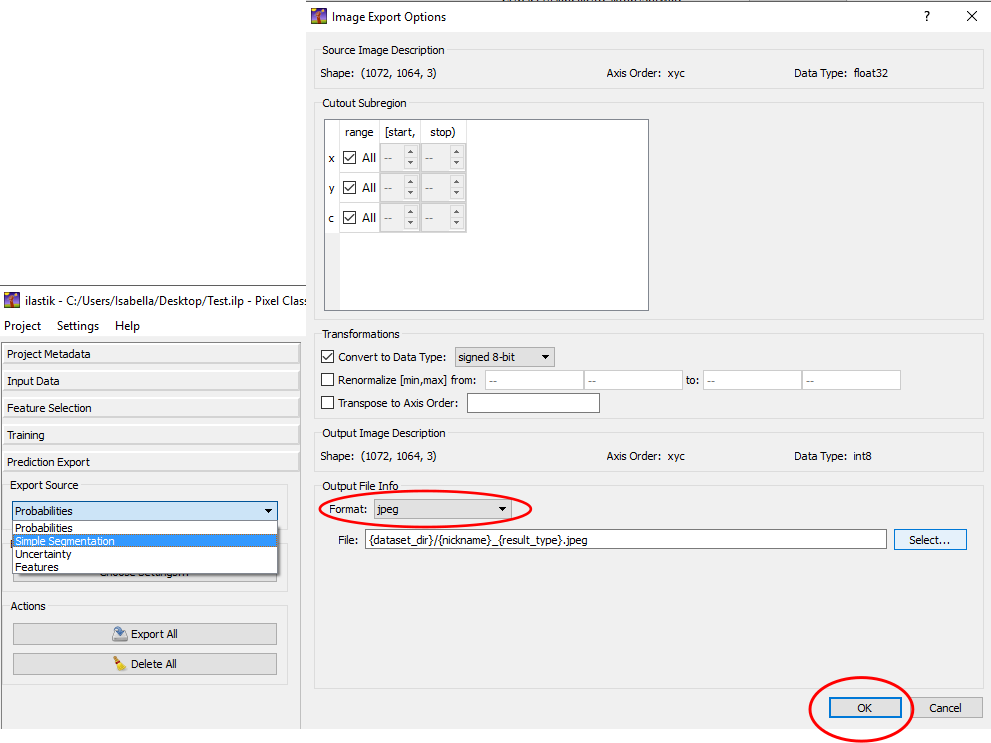
Figure 13. Image export settings after training has been completed in Ilastik
- Go to the tab ‘Prediction Export’, select by ‘Export sources’ ‘Simple segmentation’.
- Exporting images after training
- Click on ‘Export All’, the program will start with exporting the images. This can take several minutes depending on the processor and RAM module of the computer (0.9 MB/sec with an Intel® coreTM i7-4910MQ CPU @ 2.90 GHz, RAM 16 GB, for reference 50 images of each 3.08 MB take 171 sec to analyze). Exported images will contain thrips damage (red) in black and the leaf disc (green) in grey (Figure 14).

Figure 14. Exported simple segmentation of the original image. Black = thrips damage and grey = leaf disc. - Do not forget to save your project. If at a later time point you want to process additional scans of single leaf discs with the same settings, proceed to Procedure H. If you are done with processing, close the program and continue with Procedure I.
- Click on ‘Export All’, the program will start with exporting the images. This can take several minutes depending on the processor and RAM module of the computer (0.9 MB/sec with an Intel® coreTM i7-4910MQ CPU @ 2.90 GHz, RAM 16 GB, for reference 50 images of each 3.08 MB take 171 sec to analyze). Exported images will contain thrips damage (red) in black and the leaf disc (green) in grey (Figure 14).
- Processing additional images in Ilastik with the same settings
- If you want to process additional scans of single leaf discs using the same settings, make a copy of your project in the same way that you would make a copy of any office file.
- Open the copy of your project and go to the tap ‘Batch Prediction Input Selections’. Click on ‘Add new’ and import the additional images that you want to analyse.
- Go to the tab ‘Batch Prediction Output Locations’ and make sure you use the same settings for output as described in Step F2. If you keep importing images in the original project, the project becomes very large and demands heavy processing of the computer, making your computer slow. So make sure that you delete the pictures that you have imported in ‘Batch Predicition Input Selection’ before continuing with a new set of images.
- If you want to process additional scans of single leaf discs using the same settings, make a copy of your project in the same way that you would make a copy of any office file.
- Processing of simple segmentation images in ImageJ Fiji
In this step we want to extract the grey area and create images that only contain the thrips damage in black.- Open ImageJ Fiji and open the images that you want to further analyze. Click ‘File’ and select ‘Open...’ or drag your selected images from one folder directly to ImageJ Fiji.
- Open a new macro as described in Step D1, copy the following script and run the macro (only black text).imageCount = nImages
n=nImages;
for(i=0,1; i<n; i++){
setAutoThreshold("Default");
run("Threshold...");
setThreshold(0, 70);
setOption("BlackBackground", false);
run("Convert to Mask");
title=getTitle;
//Give the correct directory where you want to save the output images (marked red)
saveAs("TIFF", " X:\\...\\.....\\" + title);
close();
}
- The output images will contain the thrips damage in black (Figure 15) and are labeled the same as the original image, so make sure to save them in a separate folder.
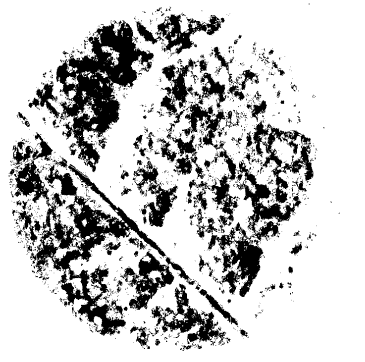
Figure 15. Image containing only thrips damage after extraction of leaf disc in ImageJ Fjij
- Open ImageJ Fiji and open the images that you want to further analyze. Click ‘File’ and select ‘Open...’ or drag your selected images from one folder directly to ImageJ Fiji.
- Calibration
Before calculating the amount of damaged surface area we determine how many pixels equal 10 mm.- Open the image that contains the calibration square or ruler in ImageJ Fiji.
- Select the strait line option and draw a line of recognizable size in your image (e.g., 10 mm on the ruler) (Figure 16).

Figure 16. Calibration in ImageJ Fiji. Selecting the straight line tool in ImageJ Fiji (A) and drawing a line (in yellow) that represents 10 mm in a scan image with a ruler (B). - Go to ‘Analyze’ and select ‘Set scale’. ImageJ Fiji produces a window that shows how many pixels equal the length of the drawn line (Figure 17, label ‘Distance in pixels:’). In this example 206.9034 pixels equal 10 mm.
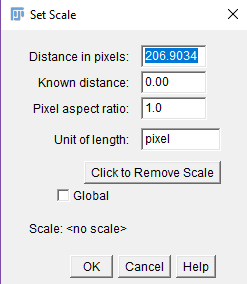
Figure 17. Window in ImageJ Fiji that shows how many pixels equal 10 mm from Figure 16B
- Open the image that contains the calibration square or ruler in ImageJ Fiji.
- Determining the amount of damaged surface area in mm2
- The damaged area can now be quantified in ImageJ Fiji. Go to ‘Process’, select ‘Batch’ and click on ‘Macro’.
- Copy the following script into the opened window. Make sure that you fill in the correct scale (marked in red text) that you obtained in Step J3.
setAutoThreshold("Default");
run("Threshold...");
setThreshold(129, 255);
run("Convert to Mask");
run("Set Scale...", "distance=#pixels known=distance in mm pixel=1 unit=mm");
run("Analyze Particles...", "show=Nothing clear include summarize") - Select the folder that contains the black and white images that were obtained in Step I3, select a different map for the output (Figure 18). ImageJ Fiji will produce copies of your analyzed images that are saved in the output folder.
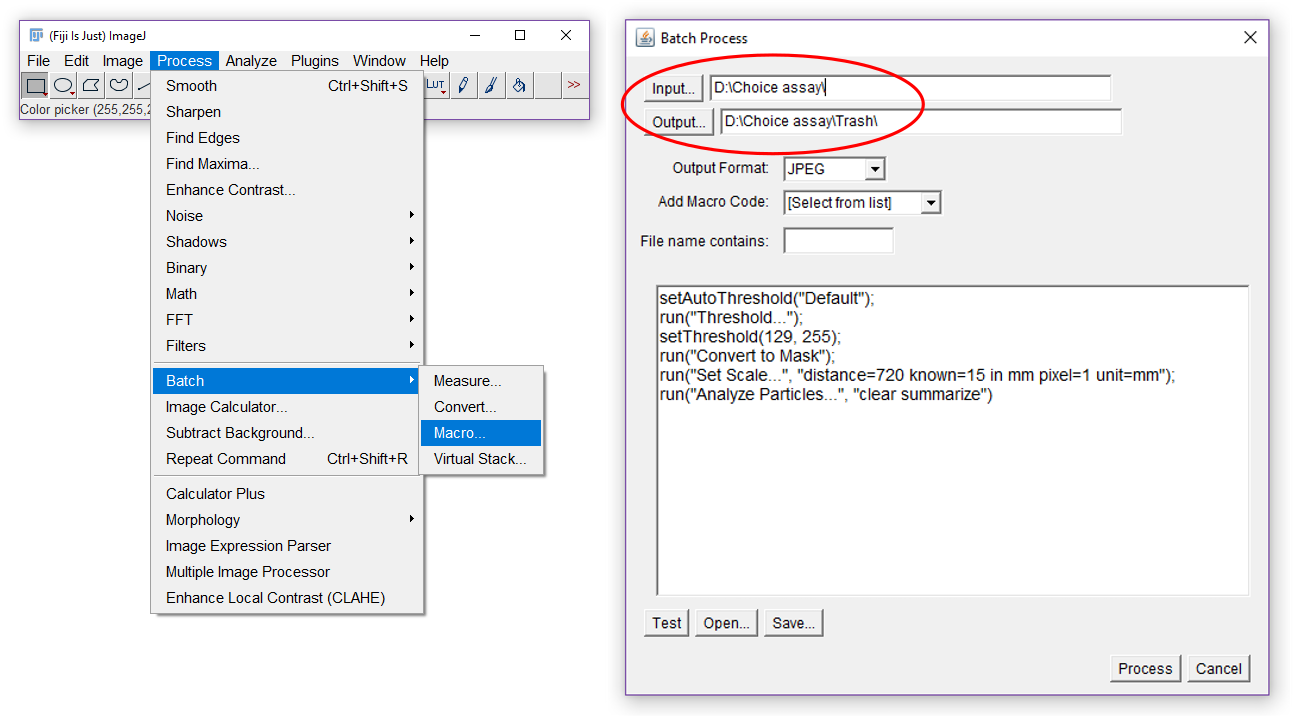
Figure 18. Batch processing in ImageJ Fjij - ImageJ Fiji will produce a summary of the measurements that you can copy to an excel file. It will contain 5 columns: ‘Slice’ (the name of the image), ‘Count’, ‘Total area’ (total thrips damage in mm2), ‘Average Size’ and ‘% Area’ (Figure 19). Copy the produced data in an excel file.
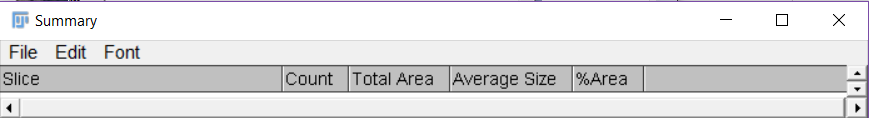
Figure 19. Output summery in ImageJ Fiji. The output shows the name of the analysed image in the column ‘Slice’ and the total thrips damage (mm2) in the column ‘Total Area’.
- The damaged area can now be quantified in ImageJ Fiji. Go to ‘Process’, select ‘Batch’ and click on ‘Macro’.
Data analysis
Before reporting the thrips damage of your leaf discs, you should correct for the average error that Ilastik makes. This average error is determined by calculating the average thrips damage on the leaf discs with no thrips (control discs). (Table 1)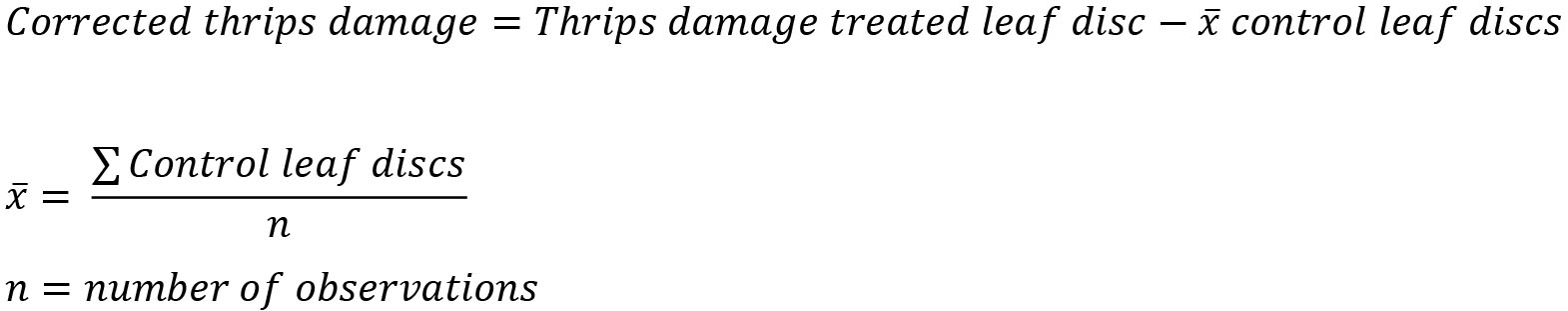
Corrected data![]()
Corrected thrips damage per leaf disc:
Image_1_T = 20 – 2 = 18
Image_1_T = 15 – 2 = 13
Image_1_T = 17 – 2 = 15
The corrected thrips damage can be used for further statistical analysis.
Table 1. Numerical example of obtained data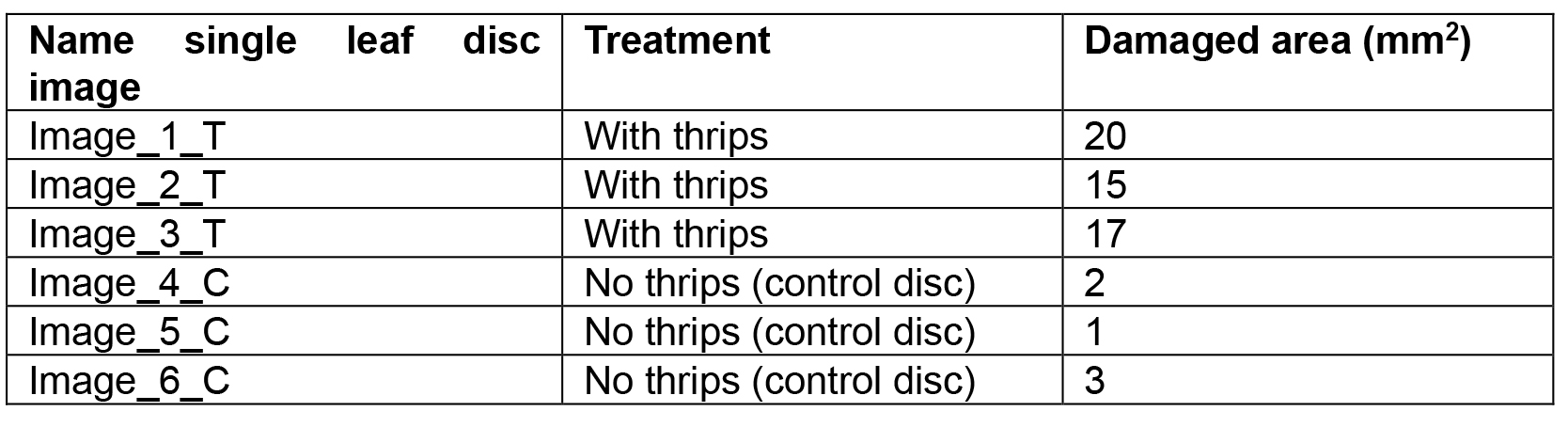
Recipes
- 1.5% liquid agar solution
Ad 7.5 g agar to 500 ml water in a 500 ml laboratory bottle.
Heat the bottle in a microwave until the agar is completely dissolved
Note: keep the cap lose on the bottle so air can escape. Otherwise the cap might blow off in the microwave due to air pressure buildup in the bottle.
Acknowledgments
This work was supported by the Stichting voor de Technische Wetenschappen (STW) which is part of the Green defense Against Pest (GAP) program, project 13552. Nicole M. van Dam gratefully acknowledges the support of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig funded by the German Research Foundation (FZT 118). This protocol has been used in Entomologia Experimentalis et Applicata (Visschers et al., 2018). The authors have declared no conflicts of interest.
References
- Bongiorno, T., Kazlow, J., Mezencev, R., Griffiths, S., Olivares-Navarrete, R., McDonald, J. F., Schwartz, Z., Boyan, B. D., McDevitt, T. C, and Sulchek, T. (2014). Mechanical stiffness as an improved single-cell indicator of osteoblastic human mesenchymal stem cell differentiation. J Biomech 47: 2197-2204.
- Fabrowski, P., Necakov, A. S., Mumbauer, S., Loeser, E., Reversi, A., Streichan, S., Briggs, J. A. G. and De Renzis, S. (2013). Tubular endocytosis drives remodelling of the apical surface during epithelial morphogenesis in Drosophila. Nat Commun 4: 2244.
- Leiss, K. A., Choi, Y. H., Abdel-Farid, I. B., Verpoorte, R. and Klinkhamer, P. G. (2009). NMR metabolomics of thrips (Frankliniella occidentalis) resistance in Senecio hybrids. J Chem Ecol 35(2): 219-229.
- Maharijaya, A., Vosman, B., Steenhuis-Broers, G., Harpenas, A., Purwito, A., Visser, R. G. F. and Voorrips, R. E. (2011). Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus). Euphytica 177: 401-410.
- Maharijaya, A., Vosman, B., Verstappen, F., Steenhuis-Broers, G., Mumm, R., Purwito, A., Visser, R. G. F. and Voorrips R. E. (2012). Resistance factors in pepper inhibit larval development of thrips (Frankliniella occidentalis). Entomol Exp Appl 145: 62-71.
- Mirnezhad, M., Romero-Gonzalez, R. R., Leiss, K. A., Choi, Y. H., Verpoorte, R. and Klinkhamer, P. G. (2010). Metabolomic analysis of host plant resistance to thrips in wild and cultivated tomatoes. Phytochem Anal 21(1): 110-117.
- Morrison, E. B. and Lindell, C. A. (2012). Birds and bats reduce insect biomass and leaf damage in tropical forest restoration sites. Ecol Appl 22: 1526-1534.
- Meyer, G. A. and Hull-Sanders, H. M. (2008). Altered patterns of growth, physiology and reproduction in invasive genotypes of Solidago gigantea (Asteraceae). Biol Invasion 10: 303-317.
- Rasband, W. S. (2011). ImageJ, US National Institutes of Health, Bethesda, Maryland, USA. https://imagej.nih.gov/ij/.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Sommer, C., Straehle, C., Köthe, U. and Hamprecht, F. A. (2011). Ilastik: Interactive learning and segmentation toolkit. 2011 IEEE International symposium on biomedical imaging: From nano to macro, Institute of Electrical and Electronics Engineers (IEEE), Yoshida Campus, Kyoto University, Kyoto, Japan, pp: 230-233.
- Visschers, I. G. S., Dam, N. M. and Peters, J. L. (2018). An objective high-throughput screening method for thrips damage quantitation using Ilastik and ImageJ. Entomologia Experimentalis et Applicata 166(6): 508-515.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Visschers, I. G. S., van Dam, N. M. and Peters, J. L. (2018). Quantification of Thrips Damage Using Ilastik and ImageJ Fiji. Bio-protocol 8(8): e2806. DOI: 10.21769/BioProtoc.2806.
Category
Plant Science > Plant immunity > Disease symptom
Plant Science > Plant physiology > Biotic stress
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










