- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Adhesion of Enteroaggregative E. coli Strains to HEK293 Cells
Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2802 Views: 6850
Reviewed by: Modesto Redrejo-RodriguezJose ThekkiniathTomas Aparicio

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Colibactin-associated Genotoxicity in HeLa Cells by In Cell Western (ICW) Using γ-H2AX as a Marker
Sophie Tronnet and Eric Oswald
Mar 20, 2018 8074 Views

Intracellular Invasion and Killing Assay to Investigate the Effects of Binge Alcohol Toxicity in Murine Alveolar Macrophages
Victor Jimenez Jr and Fernando P Monroy
Jan 20, 2019 6735 Views

Model of Chemotherapy-associated Mucositis and Oral Opportunistic Infections
Takanori Sobue [...] Anna Dongari-Bagtzoglou
Nov 5, 2019 5322 Views
Abstract
Enteroaggregative Escherichia coli (EAEC) is a recognized cause of acute diarrhea among both children and adults worldwide. EAEC strains are characterized by the presence of aggregative adherence fimbriae (AAF), which play a key role in pathogenesis by mediating attachment to the intestinal mucosa and by triggering host inflammatory responses. The aggregative adherence fimbria II (AAF/II) is the most important adherence factor of EAEC prototype strain 042 (EAEC042) to intestinal cells. Multiple receptors for AAF/II on epithelial cells have been identified including the transmembrane signaling mucin Muc1. This protocol describes a method to measure adherence of EAEC strains to HEK293 cells expressing the Muc1 glycoprotein.
Keywords: Muc1Background
EAEC is an important cause of endemic and epidemic diarrheal disease worldwide. Although most commonly associated with pediatric diarrhea in developing countries, EAEC is also linked to diarrhea in immunocompromised adults, travelers and food-borne outbreaks in the industrialized world, such as the large lethal outbreak caused by a Shiga toxin (Stx) type 2a-producing EAEC strain of serotype O104:H4 in Northern Europe in 2011 (Harrington et al., 2006; Rasko et al., 2011). EAEC pathogenesis is determined by the organism’s ability to adhere to intestinal cells, produce enterotoxins and cytotoxins, and ultimately to induce inflammation (Harrington et al., 2006). EAEC adherence to intestinal cells is mediated by AAF fimbrial adhesins (Czeczulin et al., 1997). To date, at least five variants of the AAF fimbriae have been described, all encoded in virulence plasmids ranging from 55 to 65 MDa (Jonsson et al., 2015). The AAF structure is comprised by a positively charged major subunit and a putative minor subunit at the tip of fimbrial structures (Berry et al., 2014).
The prototype EAEC strain 042 that exhibits the AAF/II variant has been shown to produce diarrhea in adult volunteers (Nataro et al., 1995). Clinical and laboratory data suggest that EAEC induces inflammatory enteritis, while studies using polarized T84 monolayers indicates that release of IL-8 is associated to the presence of AAF/II adhesin (Harrington et al., 2005). Although the importance of the adherence of EAEC to intestinal cells has been established, the cell receptors involved in the inflammatory response mediated by AAF fimbriae have not been fully characterized. Several receptors on epithelial cells have been identified for AAF/II including extracellular matrix (ECM) proteins such as fibronectin and laminin, and cytokeratin 8 (Farfan et al., 2008; Izquierdo et al., 2014). However, these receptors are localized on the basolateral side of intestinal cells. Thus, it is unlikely that these proteins play an important role during the initial infection with EAEC. Furthermore, it has been shown that fibronectin does not participate in the inflammatory response mediated by AAF/II (Yanez et al., 2016). We have recently found that EAEC also binds to the signaling Muc1 glycoprotein, and such binding is dependent on the sialylation of the protein (Boll et al., 2017). Mucins (MUC) are large (> 200 kDa) secreted and transmembrane glycoproteins with a high carbohydrate content (50-90% by weight) expressed by a variety of normal and malignant secretory epithelial cells (Corfield et al., 2001). Muc1 is a polymorphic transmembrane mucin-like protein that contains a large extracellular domain consisting of a glycosylated polypeptide made up of 30-100 tandem repeats of a 20-amino acid sequence, a transmembrane domain, and a cytoplasmic tail of 72 amino acids (Nath and Mukherjee, 2014). Muc1 is associated to numerous signaling pathways in malignant and inflammatory processes (Nath and Mukherjee, 2014). Our recent study shows that Muc1 is associated with the inflammatory response mediated by AAF/II. Moreover, EAEC 042 up-regulates epithelial Muc1 expression dependent on the presence of AAF (Boll et al., 2017).
The existence of multiple receptors for EAEC in intestinal cells complicates the identification and characterization of specific receptors in this cell lineage. The use of HEK293 cells, which differ from enterocytes, allows the evaluation of new potential receptors through transfection of these cells with plasmids encoding the candidate receptor. Likewise, binding assays performed with cells in suspension minimizes the binding of EAEC strains to non-abiotic surfaces. This protocol could be used to find out potential receptors for other enteropathogens. Here, we described in detail the method we previously used to visualize the adherence of EAEC to HEK293 cells transfected with a Muc1-encoding plasmid.
Materials and Reagents
- Cell culture flasks (Corning, catalog number: 3275 )
- 20, 200 and 1,000 μl Pipette tips (Barrier Low-Retention tips, Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 2149P-05-RT , 2069-HR and 2279 )
- 24-well plate
- Circle cover glasses (Fisher Scientific, Fisherbrand, catalog number: 12-545-80 )
- Tissue paper
- 15 ml conical culture tubes (Corning, catalog number: 430052 )
- Serological pipettes (1, 5, 10, 25, 50 ml) (Corning, catalog numbers: 4012 , 4051 , 4492 , 4251 , 4501 )
- Glass slides (Fisher Scientific, Fisherbrand, catalog number: 12-550-123 )
- Coverslip (Fisher Scientific, Fisherbrand, catalog number: 12-543C )
- Cell lines: HEK293-pcDNA3.1 and HEK293-pcDNA3.1-Muc1
HEK293 cells (ATCC, catalog number: CRL-1573 )
pcDNA3.1 (Thermo Fisher Scientific, InvitrogenTM, catalog number: V79020 )
Note: pcDNA3.1-Muc1 clone was provided by Dr. Erik P. Lillehoj (Lillehoj et al., 2002). - EAEC042 strain is available from our lab strain collection (Nataro et al., 1995)
- pGFP vector (Takara Bio, Clontech, catalog number: 632370 )
- LB medium (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10855001 )
- Glycerol (Fisher Scientific, Fisherbrand, catalog number: BP229-4 )
- Carbenicillin
- DMEM-High glucose (no phenol red), used to culture EAEC (Thermo Fisher Scientific, GibcoTM, catalog number: 31053028 )
- Phosphate buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 70011044 )
- Dulbecco’s modified Eagle’s medium (DMEM), used to culture HEK293 cells (Thermo Fisher Scientific, GibcoTM, catalog number: 11965118 )
- Fetal bovine serum, certified, heat inactivated (FBS) (Sigma-Aldrich, catalog number: F4135 )
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- G418 (Sigma-Aldrich, catalog number: G418-RO)
Manufacturer: Roche Diagnostics, catalog number: 4727878001 . - 0.5 M EDTA (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9260G )
- Methanol (Sigma-Aldrich, catalog number: 322415-1L )
- Glacial acetic acid (Fisher Scientific, Fisher Chemicals, catalog number: A38-500 )
- FITC Mouse anti-Human Muc1 (CD227) (BD, BD Biosciences, catalog number: 559774 )
- Coverslip sealant (Biotium, catalog number: 23005 )
- FM 4-64FX (Thermo Fisher Scientific, InvitrogenTM, catalog number: F34653 )
- ProLongTM Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36935 )
- 4’,6-Diamidino-2-Phenylindole (DAPI, Dilactate) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D3571 )
- Hank’s balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14175103 )
- Formalin (Sigma-Aldrich, catalog number: HT501128-4L )
Equipment
- 10, 20, 200, 1,000 μl pipettes (Pipette Pack Set, Eppendorf, catalog number: 2231300006 )
- CO2 incubator (NuAire, model: NU-4750 )
- -80 °C freezer (Thermo Fisher Scientific, Thermo ScientificTM, model: Forma Model 900 )
- Rotary shaker (ATR, model: AJ118 )
- Laminar flow hood (Thermo Fisher Scientific, Thermo ScientificTM, model: 1300 Series A2 Model 1375 )
- UV-Vis Spectrophotometer (GE Healthcare, Amersham Biosciences, model: Ultrospec 2100 Pro )
- Liquid nitrogen storage container (Worthington, model: LD35 )
- Centrifuge (Eppendorf, model: 5430 )
- Microcentrifuge (Fisher Scientific, model: accuSpinTM Micro 17R )
- Epifluorescence microscope (Olympus, model: BX51 )
Procedure
- Bacterial cell culture
- Streak out a loopful of EAEC strains (previously transformed with the pGFP plasmid and preserved at -80 °C in LB medium containing 20% glycerol) on LB agar plates containing 100 μg/ml of carbenicillin and incubate at 37 °C for 16-18 h.
- Inoculate 3-5 EAEC colonies into 5 ml LB medium and incubate at 37 °C for 16-18 h with shaking (200 rpm) using a rotary shaker. Made 1:100 dilution from this culture using fresh DMEM-High glucose (no red phenol) to induce the expression of AAF fimbria (e.g., 100 µl of bacterial culture in 9,900 µl DMEM-HG), and incubate at 37 °C with shaking (200 rpm) until reaching the mid logarithmic phase (OD600 = 0.6).
Note: DMEM-HG is known to upregulate the expression of AAF fimbriae. DMEM-HG without red phenol is used to facilitate monitoring of EAEC growth in this medium by the naked eye and spectrophotometrically. EAEC takes approximately 4 h to reach an OD600 = 0.6 when grown in DMEM-HG. - Centrifuge EAEC cultures at 5,000 x g for 10 min, wash with 10 ml 1x PBS, and resuspend in fresh DMEM (DMEM used to grow HEK293 cells) at a concentration of 1 x 108 colony forming units (CFU). We have previously determined that OD600 = 0.35 corresponds to approximately 1 x 108 EAEC CFU per ml.
- Streak out a loopful of EAEC strains (previously transformed with the pGFP plasmid and preserved at -80 °C in LB medium containing 20% glycerol) on LB agar plates containing 100 μg/ml of carbenicillin and incubate at 37 °C for 16-18 h.
- HEK293 cell culture
- Grow the human embryonic kidney cell line HEK293 (ATCC) transfected with pcDNA3.1 and pcDNA3.1-Muc1 plasmids in 75 or 150 cm2 flasks with DMEM (Gibco) supplemented with 10% FBS (Sigma-Aldrich), penicillin-streptomycin, and 200 µg/ml of G418 (Sigma-Aldrich) (to ensure maintenance of pCDNA3.1 plasmid derivatives).
- For passage and seeding purposes, wash HEK293 cells with 10 ml of sterile PBS when near to 80-90% confluency and detach using 0.35 mM EDTA in PBS as follows: add 3 ml EDTA per 75 cm2 culture flask, incubate for 15 min at 37 °C and 5% CO2 until cells have rounded and detached from the bottom, collect with 7 ml fresh medium, count cells, spin down cells for 5 min at 400 x g, and resuspend cells in fresh medium to the desired cell concentration (2 x 106/ml for binding assays).
- Grow the human embryonic kidney cell line HEK293 (ATCC) transfected with pcDNA3.1 and pcDNA3.1-Muc1 plasmids in 75 or 150 cm2 flasks with DMEM (Gibco) supplemented with 10% FBS (Sigma-Aldrich), penicillin-streptomycin, and 200 µg/ml of G418 (Sigma-Aldrich) (to ensure maintenance of pCDNA3.1 plasmid derivatives).
- Detection of Muc1 on HEK293-Muc1 cells by immunofluorescence
- Split cells onto a 24-well plate containing circle cover glasses. Pipette up and down to make sure cells are not concentrating in the center of the dish well. Grow cells to 70-90% confluency.
- Fix cells in Clark’s solution (90% methanol; 10% glacial acetic acid) for 3 h at RT.
- Remove fixative and block with 2% BSA in PBS for 30 min.
- Incubate with FITC conjugated anti Muc1 antibody (1:200 dilution) in PBS 2% BSA for 2 h at room temperature in the dark.
- Wash with PBS three times for 5 min each.
- Take out the coverslip and place it on top of a tissue paper and let it dry out. The coverslip must be facing up.
- Once the coverslip has dried out, add a drop of anti-fade with DAPI onto the coverslip.
- Mount the coverslip on the slide inverting it so the sample will be in direct contact with the slide.
- Add coverslip sealant around the coverslip to seal it on the slide. Keep the samples at -20 °C in the dark until analysis by using a fluorescent microscope.
- Split cells onto a 24-well plate containing circle cover glasses. Pipette up and down to make sure cells are not concentrating in the center of the dish well. Grow cells to 70-90% confluency.
- EAEC binding assay in HEK293 cell suspension
EAEC strains bind avidly to biotic and abiotic surfaces (such as plastic surfaces). To minimize unspecific binding to abiotic surfaces, cell-EAEC binding assays are performed in cell suspensions.
Binding assays are performed with cell suspensions in 15 ml conical tubes as follows:- Stain approximately a total of 2 x 106 cells in suspension by adding 300 µl of 5 µg/ml of FM 4-64FX (Thermo Scientific) and 4’,6’-diamidino-2-phenylindole (DAPI) for nucleus visualization for 5 min, followed by incubation with EAEC expressing GFP.
Note: DAPI can also be added at the end of the experiment as part of the anti-fade solution. HEK293 cell membranes are stained with FM 4-64FX before EAEC infection to avoid staining of bacterial membranes. If wish, FM 4-64FX staining can be skipped and proceed with the infection with fluorescent bacteria. - Remove the excess of FM 4-64FX dye on cells by adding 10 ml of HBSS to the tubes and spinning cells down at 250 x g (~1,500 rpm) for 5 min.
- Mix 1 x 106 FM 4-64FX-stained HEK293 cell derivatives expressing or not Muc1 with approximately 2 x 107 CFUs of GFP-EAEC strains to a multiplicity of infection (MOI) of 1:20 (cell:bacteria) in 1 ml of fresh DMEM medium. Incubate the cells for 1 h at 37 °C and 5% CO2.
- After 1 h incubation, wash the cells three times by adding 10 ml of HBSS buffer and by gently inverting the tube 2-3 times. Centrifuge the cells at 100 x g (~1,000 rpm) for 2 min to allow cell precipitation, but minimizing unbound bacteria sedimentation. Repeat HBSS-washes two more times.
Note: Perform washes of HEK293-Muc1 cells by gentle inversion of the tube back and forth to avoid shedding of the highly glycosylated extracellular domain of Muc1 (and disengagement of bacteria), which is not covalently attached to its transmembrane C-terminal domain. - Remove the supernatant by aspiration using a disposable transfer pipette or a vacuum system (care should be taken not to disturb or aspirate the cells at the bottom of the tube; most unbound bacteria settle at later times and lower speeds).
- Fix cells by resuspending in 100 µl of PBS-5% formalin (e.g., 50 µl PBS + 50 formalin at 5% [v:v]). Examine the cells immediately or save at 4 °C to analyze it later, preferably during the first 48 h.
- Use 10 µl of cell suspension to prepare a smear on glass slides by extending the cell suspension with a coverslip. Let smear to dry out for about 10 min, then coat smear with a drop of anti-fade-DAPI (Thermo Fisher Scientific).
- Analyze the slides by using a fluorescence microscope equipped with wavelength filters for blue (365/10 excitation/420LP emission), green (480/20 excitation/510LP emission), and red (535/30 excitation/580LP emission). You will see DNA stained in blue, cells membranes stained in red and bacteria in fluorescent green. A representative example of a binding assay with the prototype EAEC 042 strain and its isogenic fimbria mutant (042aafA) is illustrated in Figure 1.
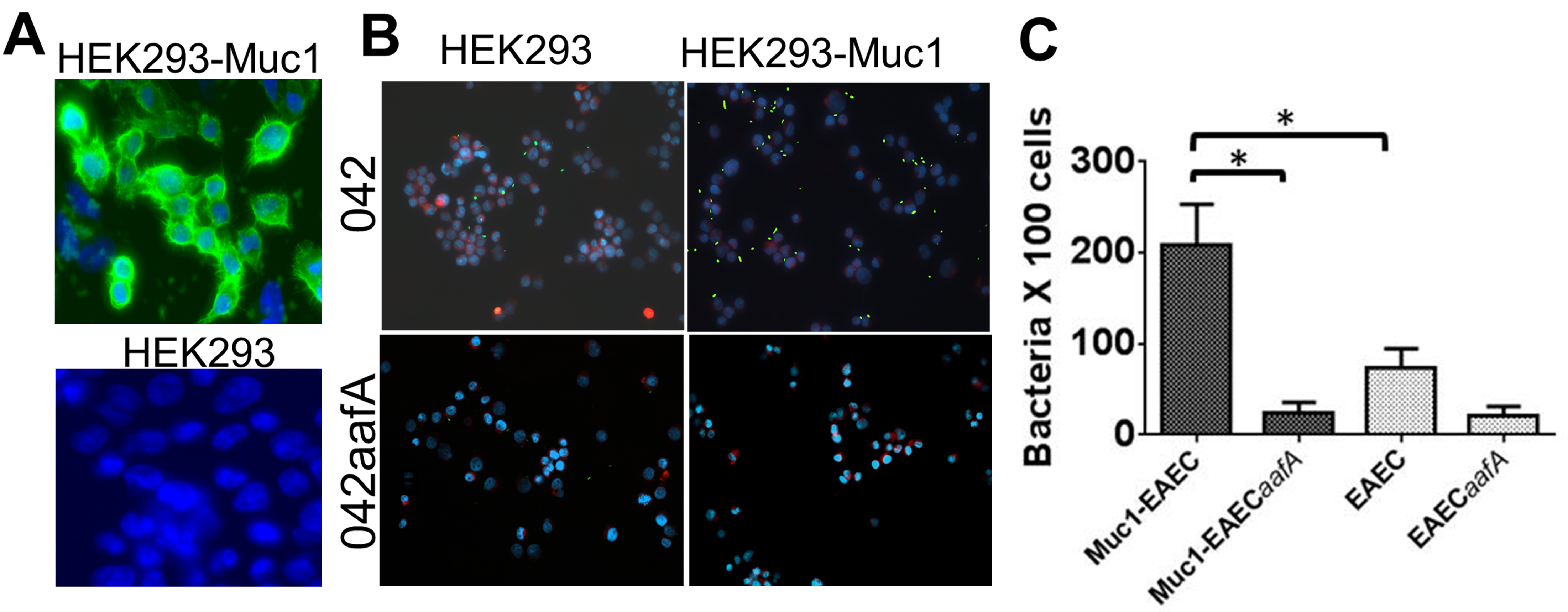
Figure 1. Binding of EAEC to HEK293-Muc1 cells. A. Production of Muc1 glycoprotein in HEK293 cells transfected with pcDNA3 (HEK293) or pcDNA3-MUC1 (HEK293-Muc1) was assessed with a FITC-conjugated monoclonal antibody to Muc1 by fluorescence microscopy (100x). B. HEK293 and HEK293-Muc1 cells were infected in suspension with EAEC strain 042 or with its isogenic fimbria mutant (042aafA), each expressing a GFP reporter and analyzed by fluorescence microscopy (40x). EAEC042 is in green, cell membranes are stained in red with FM 4-64FX, and DNA is stained in blue with DAPI. C. Bacteria adhered to cells were enumerated microscopically and data were analyzed by comparison of means in a paired t-test with significant differences P < 0.05 (*). Bars represent the means of at least three independent experiments, with the error bars indicating standard deviations.
Data analysis
EAEC adhered to HEK293 derivatives are enumerated microscopically (40x) by counting HEK293 cells and bacteria bound to cells in 20 randomized fields per coverslip from at least three independent experiments. Results are expressed as the number of bacteria per 100 cells. Data are analyzed by comparison of means in a paired t-test with significant differences when P < 0.05.
Acknowledgments
This protocol was partially reported in our previously published study (Boll et al., 2017). This work was supported by Departmental funds to FR-P (Pediatrics Department, University of Virginia). The authors declare no conflicts of interest or competing interests.
References
- Berry, A. A., Yang, Y., Pakharukova, N., Garnett, J. A., Lee, W. C., Cota, E., Marchant, J., Roy, S., Tuittila, M., Liu, B., Inman, K. G., Ruiz-Perez, F., Mandomando, I., Nataro, J. P., Zavialov, A. V. and Matthews, S. (2014). Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli. PLoS Pathog 10(9): e1004404.
- Boll, E. J., Ayala-Lujan, J., Szabady, R. L., Louissaint, C., Smith, R. Z., Krogfelt, K. A., Nataro, J. P., Ruiz-Perez, F. and McCormick, B. A. (2017). Enteroaggregative Escherichia coli adherence fimbriae drive inflammatory cell recruitment via interactions with epithelial MUC1. MBio 8(3).
- Corfield, A. P., Carroll, D., Myerscough, N. and Probert, C. S. (2001). Mucins in the gastrointestinal tract in health and disease. Front Biosci 6: D1321-1357.
- Czeczulin, J. R., Balepur, S., Hicks, S., Phillips, A., Hall, R., Kothary, M. H., Navarro-Garcia, F. and Nataro, J. P. (1997). Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun 65(10): 4135-4145.
- Farfan, M. J., Inman, K. G. and Nataro, J. P. (2008). The major pilin subunit of the AAF/II fimbriae from enteroaggregative Escherichia coli mediates binding to extracellular matrix proteins. Infect Immun 76(10): 4378-4384.
- Harrington, S. M., Dudley, E. G. and Nataro, J. P. (2006). Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett 254(1): 12-18.
- Harrington, S. M., Strauman, M. C., Abe, C. M. and Nataro, J. P. (2005). Aggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with enteroaggregative Escherichia coli. Cell Microbiol 7(11): 1565-1578.
- Izquierdo, M., Navarro-Garcia, F., Nava-Acosta, R., Nataro, J. P., Ruiz-Perez, F. and Farfan, M. J. (2014). Identification of cell surface-exposed proteins involved in the fimbria-mediated adherence of enteroaggregative Escherichia coli to intestinal cells. Infect Immun 82(4): 1719-1724.
- Jonsson, R., Struve, C., Boisen, N., Mateiu, R. V., Santiago, A. E., Jenssen, H., Nataro, J. P. and Krogfelt, K. A. (2015). Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect Immun 83(4): 1396-1405.
- Lillehoj, E. P., Kim, B. T. and Kim, K. C. (2002). Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am J Physiol Lung Cell Mol Physiol 282(4): L751-756.
- Nataro, J. P., Deng, Y., Cookson, S., Cravioto, A., Savarino, S. J., Guers, L. D., Levine, M. M. and Tacket, C. O. (1995). Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis 171(2): 465-468.
- Nath, S. and Mukherjee, P. (2014). MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 20(6): 332-342.
- Rasko, D. A., Webster, D. R., Sahl, J. W., Bashir, A., Boisen, N., Scheutz, F., Paxinos, E. E., Sebra, R., Chin, C. S., Iliopoulos, D., Klammer, A., Peluso, P., Lee, L., Kislyuk, A. O., Bullard, J., Kasarskis, A., Wang, S., Eid, J., Rank, D., Redman, J. C., Steyert, S. R., Frimodt-Moller, J., Struve, C., Petersen, A. M., Krogfelt, K. A., Nataro, J. P., Schadt, E. E. and Waldor, M. K. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365(8): 709-717.
- Yanez, D., Izquierdo, M., Ruiz-Perez, F., Nataro, J. P., Giron, J. A., Vidal, R. M. and Farfan, M. J. (2016). The role of fibronectin in the adherence and inflammatory response induced by enteroaggregative Escherichia coli on epithelial cells. Front Cell Infect Microbiol 6: 166.
- Stain approximately a total of 2 x 106 cells in suspension by adding 300 µl of 5 µg/ml of FM 4-64FX (Thermo Scientific) and 4’,6’-diamidino-2-phenylindole (DAPI) for nucleus visualization for 5 min, followed by incubation with EAEC expressing GFP.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ayala-Lujan, J. L. and Ruiz-Perez, F. (2018). Adhesion of Enteroaggregative E. coli Strains to HEK293 Cells. Bio-protocol 8(8): e2802. DOI: 10.21769/BioProtoc.2802.
Category
Microbiology > Microbe-host interactions > In vitro model > Cell line
Immunology > Mucosal immunology > Epithelium
Cell Biology > Cell-based analysis > Cell adhesion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








