- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Commensal Escherichia coli Strains from Feces of Healthy Laboratory Mice or Rats
Published: Vol 8, Iss 6, Mar 20, 2018 DOI: 10.21769/BioProtoc.2780 Views: 10906
Reviewed by: David CisnerosWolf Dieter RötherAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2896 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2089 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1657 Views
Abstract
The colonization abundance of commensal E. coli in the gastrointestinal tract of healthy laboratory mice and rats ranges from 104 to 106 CFU/g feces. Although very well characterized, the family that E. coli belongs to has a very homogeneous 16S rRNA gene sequence, making the identification from 16S rRNA sequencing difficult. This protocol provides a procedure of isolating and identifying commensal E. coli strains from a healthy laboratory mouse or rat feces. The method can be applied to isolate commensal E. coli from other laboratory rodent strains.
Keywords: CommensalBackground
Escherichia coli is a Gram-negative, facultative anaerobe which constitutes only a minor fraction of the vertebrate gut microbiota, but plays a key role in microbial interaction, immune modulation and metabolic functionalities (Tenaillon et al., 2010). Being one of the best-characterized model microorganisms, commensal E. coli strains have been increasingly studied to unravel the mechanisms through which gut commensal microbes adapt to the unique niche and impact host physiology. However, the high homology among different strains raises difficulties in identification and characterization of commensal E. coli based on a 16S rRNA sequencing approach. Thanks to the development of next-generation sequencing techniques and large-scale analyses of whole genomes, we are able to identify commensal E. coli strains isolated from the gastrointestinal tract of different hosts according to the presence of virulence genes in the genome. In this protocol, we show an approach to isolate and identify commensal E. coli strains from a laboratory mouse or rat using selective culture media and whole genome sequencing. However, it should be noted that the presence of commensal E. coli in laboratory animals depends on the vendor and environmental conditions of the facility.
Materials and Reagents
- Gloves and masks (KCWW, Kimberly-Clark, catalog number: 52817 ; Cardinal Health, Insta-Gard, catalog number: AT7511-WE )
- 1.5 ml centrifuge tube (sterile) (Fisher Scientific, catalog number: 05-408-129 )
- Wide bore tips, 0-200 μl (Corning, Axygen®, catalog number: T-1005-WB-C )
- Thin-wall PCR tubes (Fisher Scientific, catalog number: 14-230-225 )
- Tips, 0.1-10 μl, 0.1-1 ml (sterile) (Fisher Scientific, catalog numbers: 02-707-474 ; 02-707-480 )
- Cell spreader (Fisher Scientific, catalog number: 08-100-11 )
- Petri dishes (100 x 15 mm) (Fisher Scientific, catalog number: FB0875713 )
- 15 ml conical sterile polypropylene centrifuge tubes (Thermo Fisher Scientific, NuncTM, catalog number: 339650 )
- Sterile 0.22 μm filter (Corning, catalog number: 431219 )
- 1 ml syringe (BD, catalog number: 309659 )
- A healthy NIH Swiss mouse (Harlan Laboratories Inc., Indianapolis, IN) and Sprague-Dawley (SD) rat (Charles River Canada, St. Constant, QC)
- 70% ethanol diluted from 100% ethanol (Commercial Alcohols, catalog number: P016EAAN )
- Agarose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 16500500 )
- Tris-Acetate-EDTA (TAE) (50x stock) (Fisher Scientific, catalog number: BP13321 )
- SYBR Safe DNA gel stain (Thermo Fisher Scientific, InvitrogenTM, catalog number: S33102 )
- 6x DNA loading dye (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0611 )
- 1 kb Plus DNA ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM1331 )
- GeneJET gel extraction and DNA cleanup micro kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K0832 )
- PureLink genomic DNA mini kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: K182001 )
- Nextera XT DNA library preparation kit (Illumina, catalog number: FC-131-1096 )
- Nextera XT DNA library preparation index kit (Illumina, catalog number: FC-131-1002 )
- Nextera XT DNA library preparation kit (Illumina, catalog number: FC-131-1096 )
- Nextera XT DNA library preparation index kit (Illumina, catalog number: FC-131-1002 )
- QubitTM 1x dsDNA HS assay kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q33230 )
- PhiX control kit (Illumina, catalog number: FC-110-3001 )
- MiSeq reagent kit V3 (Illumina, catalog number: MS-102-3003 )
- Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: S373-500 )
- Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number: P285-500 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-500 )
- Potassium chloride (KCl) (Fisher Scientific, catalog number: P217-500 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144-500LB )
- MacConkey agar (BD, catalog number: 212123 )
- Luria-Bertani (LB) broth (Sigma-Aldrich, catalog number: L3022 )
- Glycerol (Fisher Scientific, catalog number: BP229-1 )
- DNA Taq polymerase with 50 mM MgCl2 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10342020 )
- Oligo primers 27F/1492R (Weisburg et al., 1991)
27F: 5’-AGAGTTTGATCMTGGCTCAG-3’
1492R: 5’-TACGGYTACCTTGTTACGACTT-3’ - Deoxynucleotide triphosphates (dNTPs) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10297-018 )
- PCR grade water (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9932 )
- Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: SS266-1 )
- 1x phosphate buffered saline (PBS) (pH 7.4) (see Recipes)
- MacConkey agar (see Recipes)
- LB broth (see Recipes)
- Glycerol stock of bacterial isolates (see Recipes)
- PCR reaction mix (see Recipes)
- 0.1 N NaOH (see Recipes)
Equipment
- Forceps (sterilized by autoclave)
- Vortex (Fisher Scientific, catalog number: 02215365 )
- Pipettes [e.g., P1000 (Eppendorf, catalog number: 3120000062 ), P200 (Eppendorf, catalog number: 3120000054 ), P10 (Eppendorf, catalog number: 3120000020 )]
- Thermal cycler (Thermo Fisher Scientific, Applied Biosystems, model: GeneAMP PCR System 9700 )
- Microwave (RCA, catalog number: RMW733 )
- Gel electrophoresis system (Fisher Scientific, model: FB300 )
- UV transilluminator with photo documentation (Azure Biosystems, model: Azure c200 )
- Incubator (37 °C) (Thermo Fisher Scientific, Thermo Scientific, model: Model 370 )
- Shaking incubator (37 °C) (Eppendorf, New BrunswickTM, model: I26 )
- QubitTM 3.0 fluorometer (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q33216 )
- QubitTM assay tubes (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32856 )
- Illumina MiSeq instrument (Illumina, model: MiSeqTM System, catalog number: SY-410-1003 )
- Autoclave (Beta Star Life Science Equipment, model: C2002BS )
- -80 °C freezer (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM 900 Series , 989)
Procedure
The protocol for isolating and identifying the gut commensal E. coli strains from laboratory mouse or rat feces includes the steps for fecal collection, bacterial culture and colony characterization (Figure 1).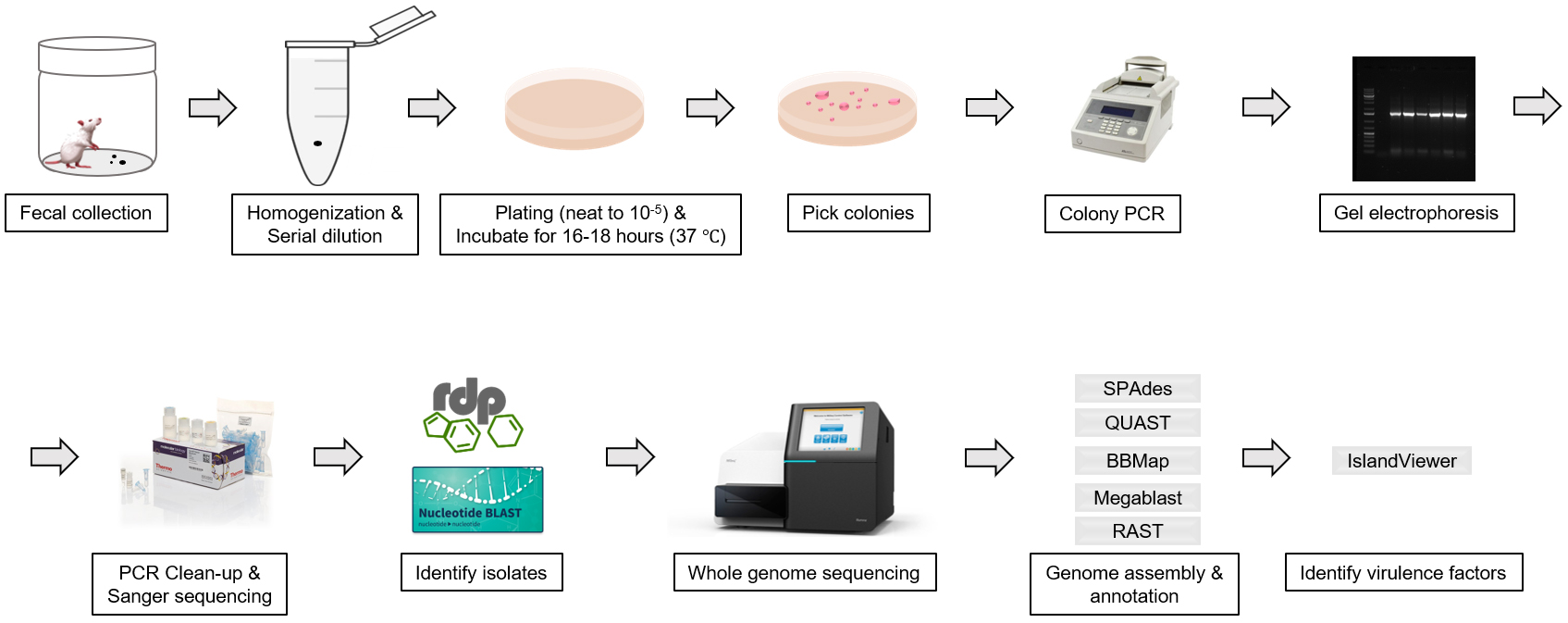
Figure 1. Workflow scheme for the isolation and identification of commensal E. coli from mouse or rat feces
- Fecal collection
Note: Personal protection equipment including gloves and masks are required.- Put a healthy NIH Swiss mouse/SD rat into a clean cage and wait for defecation.
- Use sterile forceps to transfer one fresh fecal pellet into a 1.5 ml centrifuge tube preloaded with 1 ml of 1x PBS.
Note: Recommended weight of one fresh fecal pellet: mouse, 30-50 mg; rat, 80-100 mg. - Put the tubes with fecal pellet on the ice and immediately transfer them back to the lab to perform serial dilution and plating.
- Put a healthy NIH Swiss mouse/SD rat into a clean cage and wait for defecation.
- Serial dilution and plating
Note: Wear gloves and use 70% ethanol as a disinfectant for cleaning the surfaces of working bench.- Homogenize the fecal pellet thoroughly by vortexing for 30 sec at the maximum speed.
- Add 900 μl of 1x PBS into four 1.5 ml centrifuge tubes. Perform 1:10 serial dilution from the fecal samples using the wide bore tips.
- Use the cell spreaders to plate 100 μl of each dilution onto MacConkey agar plate (10-1 to 10-5).
- Incubate the plates aerobically at 37 °C for 16-18 h.
- Homogenize the fecal pellet thoroughly by vortexing for 30 sec at the maximum speed.
- Colony PCR for E. coli identification by Sanger sequencing using 16S rRNA gene primers
- MacConkey agar is a selective and differential medium for the isolation of coliform organisms. The potential E. coli colonies appear as red and round shaped colonies due to the capability to ferment lactose (Figure 2). Select potential colonies for the following PCR amplification.

Figure 2. Colony morphology of E. coli isolated from rat feces after growth on MacConkey agar. The potential E. coli colonies appear as red with a round shape. The colonies pointed with black arrows are considered as negative colonies. - Add 50 μl of PCR reaction mix (see Recipes) into each PCR tube.
- Use P10 tips to pick a small amount of the single colony and swirl in the PCR reaction. Mix the colonies and PCR reaction by pipetting up and down, and then discard the tips.
- Complete the reaction in a thermal cycler following the PCR program (see Notes).
- Prepare 40 ml of 1% agarose gel by dissolving 0.4 g agarose in 1x TAE and heating (microwave). Add 4 μl of SYBR Safe DNA gel stain into the gel and mix thoroughly. Cast the agarose gel in the apparatus and wait until the gel is solidified. Mix 5 μl of PCR reaction with 1 μl 6x DNA loading dye and apply the samples as well as the marker (2 μl, 1 kb Plus DNA ladder) to the gel. Run the electrophoresis for 30 min at 100 V. Take a picture to check the amplification using a UV transilluminator. Compare the size of the PCR product (about 1,465 bp) to the marker (Figure 3).
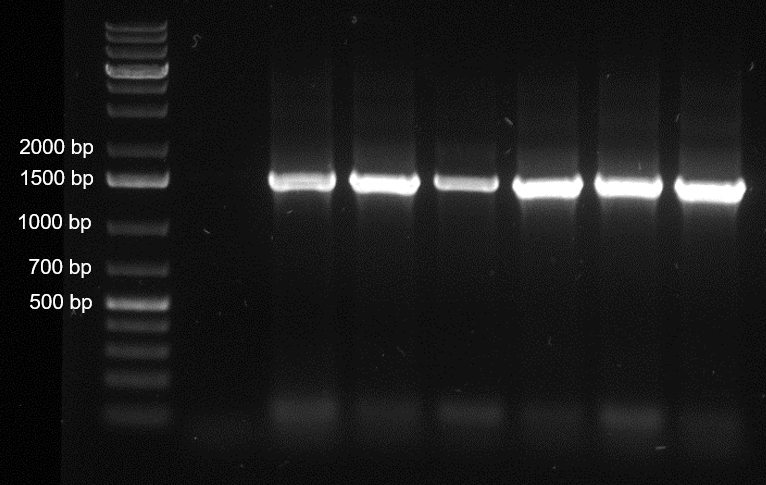
Figure 3. Agarose gel electrophoresis image of the 16S rRNA genes amplified from six bacterial isolates. Lane 1 indicates the negative control and the lane 2 to 7 indicate the PCR products amplified the 16S rRNA gene of the selected bacterial isolates. The size of the PCR product is around 1,465 bp. - PCR clean-up is performed using a GeneJET gel extraction and DNA cleanup kit following the manufacturer’s instruction. The remaining PCR mix after the gel electrophoresis is used for this step. With the adjusted concentrations, the cleaned-up samples are further sequenced by Sanger sequencing using the 16S rRNA gene primers.
- The 16S rRNA gene sequences are searched against the Ribosomal Database Project (RDP, released 11.4; http://rdp.cme.msu.edu/) and the NCBI nucleotide database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (see Data analysis).
- The isolates that share higher than 98% 16S rRNA sequence identity with the type strain of E. coli are selected and cultured in LB broth (see Recipes) overnight in a shaking incubator (37 °C) to make glycerol stocks (25% glycerol, see Recipes) for preservation.
- MacConkey agar is a selective and differential medium for the isolation of coliform organisms. The potential E. coli colonies appear as red and round shaped colonies due to the capability to ferment lactose (Figure 2). Select potential colonies for the following PCR amplification.
- Whole genome sequences and annotation
- The selected isolates are cultured overnight in 5 ml LB broth for genomic DNA extraction using a PureLink Genomic DNA Mini Kit following the manufacturer’s instruction. The LB culture from the last Step C8 can be utilized for DNA extraction.
- The whole genome sequencing is performed on an Illumina MiSeq platform. The isolated genomic DNA is fragmented to generate libraries using a Nextera XT DNA library preparation kit according to the manufacturer’s instruction.
- The concentrations of generated libraries are quantified by a QubitTM 3.0 fluorometer using a QubitTM 1x dsDNA HS assay kit according to the manufacturer’s instruction. The quantified libraries are further normalized to 2 nM and pooled following the protocol of the Nextera XT DNA library preparation kit.
- Denature the pooled libraries using 0.1 N NaOH and mix with 5% PhiX genomic DNA as a positive control.
- The sequencing of denatured libraries is performed on an Illumina MiSeq instrument with 2 x 300 bp reads generated, using a MiSeq reagent V3 sequencing-by-synthesis kit.
- The draft genome is assembled with the SPAdes assembler (Bankevich et al., 2012). Genome assemblies are evaluated by Quality Assessment Tool for Genome Assemblies (QUAST) (Gurevich et al., 2013). The tool of BBMap (Bushnell, 2014) is used to map the raw reads back to the contigs produced by SPAdes to obtain the information about the coverage for contigs. The algorithm of Megablast (Zhang et al., 2000) is applied to blast the contigs against the reference bacterial genomes obtained from NBCI. Rapid Annotations using Subsystems Technology (RAST) (Aziz et al., 2008) is used for genome annotation.
- IslandViewer (Dhillon et al., 2015) is used to predict toxin related virulence in the whole genome of the E. coli isolate. The genomes of the isolates submitted to IslandViewer are in the format of GENBANK. The isolates without identified hits of toxin virulence factor (VF)-related genes in the genome are considered to be commensal E. coli isolates.
- The selected isolates are cultured overnight in 5 ml LB broth for genomic DNA extraction using a PureLink Genomic DNA Mini Kit following the manufacturer’s instruction. The LB culture from the last Step C8 can be utilized for DNA extraction.
Data analysis
After colony PCR and Sanger sequencing, the 16S rRNA sequences of bacterial isolates are aligned against the RDP and NCBI nucleotide database. The tools of Seqmatch and Classifier within the RDP database are used for assigning taxonomy. High-quality sequences of the type strains (the size ≥ 1,200 bp) are chosen in the settings of Seqmatch. The ‘16S ribosomal RNA sequences’ database (Bacteria and Archaea) is used as the reference database with default settings when searching against the NCBI database.
Notes
- Thermal cycler condition
Initial denaturation for 10 min at 94 °C
35 cycles of 30 sec at 94 °C, 30 sec at 58 °C, and 1 min 40 sec at 72 °C
Final extension for 7 min at 72 °C
Hold at 4 °C
Recipes
- 1x phosphate buffer saline (PBS) (pH 7.4) (1 L)
10 mM Na2HPO4
1.8 mM KH2PO4
137 mM NaCl
2.7 mM KCl
Adjust to pH 7.4 with HCl and sterilize by filter or autoclave before use - MacConkey agar
Suspend 50 g of the powder in 1 L of ddH2O. Mix thoroughly and boil for 1 min to completely dissolve the powder. Autoclave at 121 °C for 15 min. Cool down and dispense approximately 20 ml per Petri dish (100 x 15 mm in diameter). Store at 4 °C for up to a month - LB broth
Stir to suspend 25 g of the powder in 1 L of ddH2O. Autoclave for 15 min at 121 °C to sterilize. Store at 4 °C for up to a month - Glycerol stock of bacterial isolates (in 25% glycerol)
- Prepare 100 ml of 50% (v/v) glycerol
Glycerol (100%) 50 ml
Add ddH2O to 100 ml
Autoclave to sterilize - Make glycerol stock from the broth culture of bacterial isolates
Use sterile pipet tips, pipet 500 μl of bacterial broth culture into a sterile centrifuge tube
Add 500 μl of 50% autoclaved glycerol. Mix the solution by vortex and place the tubes into the -80 °C freezer
- Prepare 100 ml of 50% (v/v) glycerol
- PCR reaction mix
5 μl of 10x PCR buffer (1x final concentration)
0.5 μl of 1 U/μl Taq polymerase
2 μl of 50 mM MgCl2
2 μl of 10 μM Oligo primer 27F
2 μl of 10 μM Oligo primer 1492R
2 μl of 10 mM dNTP mix
Add up to 50 μl of total volume with PCR grade water - 0.1 N NaOH solution
Make a 1:10 dilution of 1 N NaOH (Fisher Scientific) using nuclease-free water. For example: In a sterile 15 ml centrifuge tube, add 9 ml of nuclease-free water into 1 ml of 1 N NaOH solution. Mix the solution by vortex and sterilize by filtration through a 0.22 μm filter with a syringe
Acknowledgments
This research was supported by a Natural Sciences and Engineering Research Council Discovery grant held by B.P.W. T.J. was supported by a Graduate Student Scholarship from Alberta Innovates-Technology Futures, Alberta, Canada. B.P.W. is supported by the Canada Research Chair Program. This protocol was adapted from Ju et al., 2017. The authors have no conflict of interest to declare.
References
- Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., Kubal, M., Meyer, F., Olsen, G. J., Olson, R., Osterman, A. L., Overbeek, R. A., McNeil, L. K., Paarmann, D., Paczian, T., Parrello, B., Pusch, G. D., Reich, C., Stevens, R., Vassieva, O., Vonstein, V., Wilke, A. and Zagnitko, O. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75.
- Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A. and Pevzner, P. A. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5): 455-477.
- Bushnell, B. (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner. United States.
- Dhillon, B. K., Laird, M. R., Shay, J. A., Winsor, G. L., Lo, R., Nizam, F., Pereira, S. K., Waglechner, N., McArthur, A. G., Langille, M. G. and Brinkman, F. S. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 43(W1): W104-108.
- Gurevich, A., Saveliev, V., Vyahhi, N. and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8): 1072-1075.
- Ju, T., Shoblak, Y., Gao, Y., Yang, K., Fouhse, J., Finlay, B. B., So, Y. W., Stothard, P. and Willing, B. P. (2017). Initial gut microbial composition as a key factor driving host response to antibiotic treatment, as exemplified by the presence or absence of commensal Escherichia coli. Appl Environ Microbiol 83.
- Tenaillon, O., Skurnik, D., Picard, B. and Denamur, E. (2010). The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8(3): 207-217.
- Weisburg, W. G., Barns, S. M., Pelletier, D. A. and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2): 697-703.
- Zhang, Z., Schwartz, S., Wagner, L. and Miller, W. (2000). A greedy algorithm for aligning DNA sequences. J Comput Biol 7(1-2): 203-214.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ju, T. and Willing, B. P. (2018). Isolation of Commensal Escherichia coli Strains from Feces of Healthy Laboratory Mice or Rats. Bio-protocol 8(6): e2780. DOI: 10.21769/BioProtoc.2780.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial cell biology > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











