- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Plant Cell Death by Electrolyte Leakage Assay
Published: Vol 8, Iss 5, Mar 5, 2018 DOI: 10.21769/BioProtoc.2758 Views: 24451
Reviewed by: Andrea PuharBin TianAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1343 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 408 Views
Abstract
We describe a protocol to measure the electrolyte leakage from plant tissues, resulting from loss of cell membrane integrity, which is a common definition of cell death. This simple protocol is designed to measure the electrolyte leakage from a tissue sample over a time course, so that the extent of cell death in the tissue can be monitored dynamically. In addition, it is easy to handle many tissue samples in parallel, which allows a high level of biological replication. Although the protocol is exemplified by cell death in Arabidopsis in response to pathogen challenge, it is easily applicable to other types of plant cell death.
Keywords: PlantsBackground
When a cell dies and loses the integrity of the cell membrane, electrolytes, such as K+ ions, leak out of the cell. Thus, we can use the amount of electrolytes leaked from a tissue as a proxy for the extent of cell death in the tissue. A simple way to quantify such electrolytes leaked from a tissue is to measure the increase in electrolytic conductivity of water that contains the tissue with dying cells. This electrolyte leakage assay has been applied to plant tissues to assess the relative quantity of cells that died in response to biotic and abiotic stresses, such as pathogen challenge, insect herbivory, wounding, UV radiation, oxidative stress, salinity, drought, cold and heat stress (Demidchik et al., 2014).
The original method was designed to measure the conductivity of the aqueous bathing solution containing plant tissues before and after boiling it, in which the conductivity after boiling was used to normalize tissue size differences (Whitlow et al., 1992). Here we describe a procedure to dynamically monitor electrolyte leakage from leaf disks by measuring at multiple time points the electrolytic conductivity of water on which the leaf disks float in a 12-well plate. It is reasonable to assume that the total amount of electrolytes from tissue samples of the same size, such as disks of the same area punched out from leaves of a similar developmental stage, is comparable and that it is not necessary to measure the electrolytic conductivity after boiling the tissues. We inoculated leaves with the bacterial pathogen, Pseudomonas syringae pv. tomato DC3000 pVSP61-avrRpt2 (Pto DC3000 avrRpt2) in order to trigger a type of programmed cell death, known as hypersensitive cell death. The protocol presented here has been applied to our studies (Igarashi et al., 2013; Bethke et al., 2016; Hatsugai et al., 2016; Hatsugai et al., 2017) and could also be used to quantify plant cell death triggered by any other stimuli. If detailed comparisons of the time courses of the electrolyte leakage are needed, fitting polynomial regression to the time courses is possible (Van Poecke et al., 2007; Qi et al., 2010), as it is easy to obtain electrolytic conductivity measurements at many time points with many replicates.
Materials and Reagents
- 2 ml microcentrifuge tubes (Fisher Scientific, catalog number: 05-408-138 )
- Sterilized tubes for liquid bacterial cultures (Evergreen Scientific, catalog number: 222-2094-050 )
- Disposable 1-ml needleless syringes for bacterial inoculation (BD, catalog number: 309659 )
- 12-well cell culture plates (flat bottom with lid) (Corning, Costar®, catalog number: 3513 )
- 1-200 μl Pipette tips (Sorenson BioScience, catalog number: 3211 )
- 50-1,250 μl pipette tips (Sorenson BioScience, catalog number: 3205 )
- Arabidopsis thaliana accession Col-0 (Figure 1A)
Note: Arabidopsis thaliana accession Col-0 carries the R gene RPS2, which confers resistance to Pto DC3000 avrRpt2 (Bent et al., 1994; Mindrinos et al., 1994). - Pseudomonas syringae pv. tomato DC3000 pVSP61-avrRpt2 (Pto DC3000 avrRpt2)
Note: Pto DC3000 avrRpt2 delivers the AvrRpt2 effector into plant cells, thereby inducing hypersensitive cell death in Arabidopsis Col-0. - Sterilized ultrapure water (e.g., Milli-Q)
- Conductivity standard solution 1.41 mS/cm (HORIBA, model: Y071L, catalog number: 514-22 )
- Antibiotics
- Bacto proteose peptone No. 3 (BD, catalog number: 211693 )
- Glycerol (Fisher Scientific, catalog number: G33-500 )
- Dibasic potassium phosphate (K2HPO4) (Fisher Scientific, catalog number: BP363-500 )
- Bacto agar (BD, catalog number: 214010 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A508-P500 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391 )
- King’s B liquid medium (see Recipes)
Equipment
- Walk-in Arabidopsis growth chamber (22 °C, 70% relative humidity, and 12-h/12-h day/night photoperiod) (Conviron, model: BDR40 )
- Tissue culture roller rotator drum for bacterial culture at 28 °C (New Brunswick Scientific, model TC-7 )
- Centrifuge (Eppendorf, model: 5415 D )
- Spectrophotometer to determine the density of bacterial culture (Beckman Coulter, model: DU-800 )
- Cork borer (size 4, diameter = 7.5 mm)
- Electrolytic conductivity meter (HORIBA, model: B-173 )
- Single-channel micropipettor (Eppendorf, 20-200 μl and 100-1,000 μl)
- Autoclave
Software
- Microsoft Excel
Procedure
- Inoculation of Arabidopsis leaves with Pto DC3000 avrRpt2
- Culture the bacterial strain in 2 ml King’s B liquid medium supplemented with 50 µg/ml kanamycin and 50 µg/ml rifampicin at 28 °C for 16-18 h.
Note: The bacterial culture used should be in a late log phase of growth (OD600 = 1.5 - 2.0). - Transfer 1 ml of the liquid culture to a 2-ml microcentrifuge tube.
- Harvest bacteria by centrifugation at 3,000 x g for 5 min at room temperature.
- Remove supernatant and then suspend bacteria in 1 ml sterilized ultrapure water.
- Repeat Steps A3-A4 one more time.
- Adjust the optical density of the bacterial suspension with sterilized ultrapure water to OD600 = 0.1 (measured by a spectrophotometer).
- Pressure-infiltrate the density-adjusted bacterial suspension (Katagiri et al., 2002) into two leaves per plant in a walk-in Arabidopsis growth chamber.
Note: Gently pressure-infiltrate the bacterial suspension through the stomatal openings on the abaxial side of the leaf using a 1-ml needleless syringe. The infiltrated area of a leaf is readily visible as the appearance of the infiltrated area turns dark green due to flooding of the intercellular space of the leaf with bacterial suspension.
- Culture the bacterial strain in 2 ml King’s B liquid medium supplemented with 50 µg/ml kanamycin and 50 µg/ml rifampicin at 28 °C for 16-18 h.
- Measurement of electrolytic conductivity
- Cut two leaf disks (7.5 mm diameter) from one plant (one disk per leaf) using a cork borer on paper towels (Figure 1B). Leaf disks should be cut out before the infiltrated bacterial suspension from Step A7 dries out from the intercellular space of the leaves, which occurs approximately 20 min after infiltration.
Note: We typically use 4-weeks old plants grown under the growth conditions described in Equipment section and select the 7th and 8th leaves of each plant to obtain leaves of comparable developmental stages. Selecting leaves in similar developmental stages is important not only to have a similar amount of tissue for each leaf disk but also to have a similar response to the pathogen. - Float two leaf disks (adaxial surface down) from a single plant on 2 ml of sterilized ultrapure water in one well of a 12-well plate, immediately after the leaf disks are cut out in Step B1 (Figure 1C). Use as many wells and 12-well plates as needed according to the number of plants to be assayed.
- Cover the plate with the lid and incubate it for 30 min in a walk-in Arabidopsis growth chamber.
- Replace the water in the well with 2 ml of fresh sterilized ultrapure water.
Note: Steps B3 and B4 are to remove electrolytes initially leaked from the damaged cells on the edges of the leaf disks. - Incubate the plate in a walk-in Arabidopsis growth chamber.
- Calibrate the electrolytic conductivity meter before the first use with the conductivity standard solution to 1.41 mS/cm.
- Wash the sensor of the electrolytic conductivity meter with sterilized ultrapure water.
- Sample 100 µl of the water per well at the determined time point and measure its conductivity using the sensor of an electrical conductivity meter (Figure 1D).
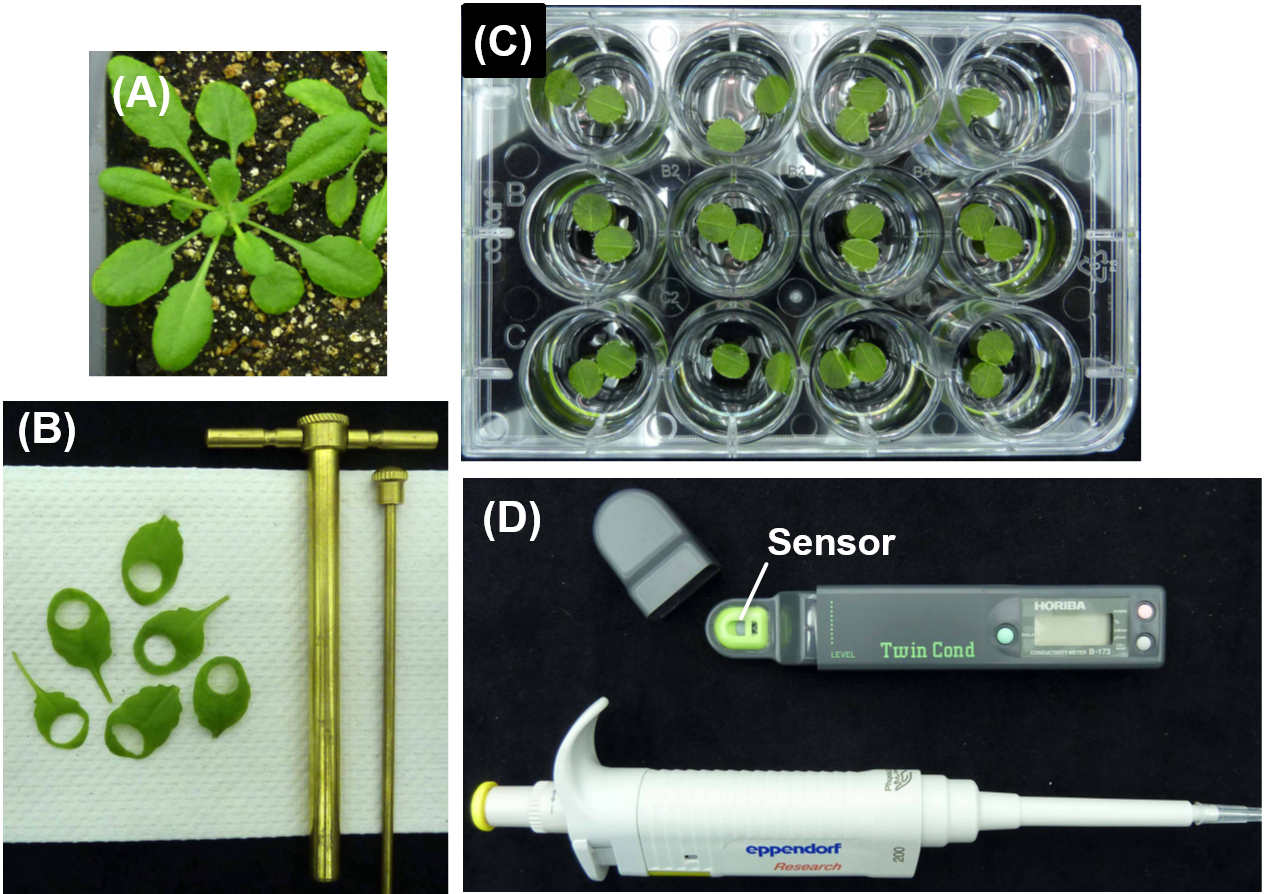
Figure 1. Experimental procedure to measure the electrolytic leakage from Arabidopsis leaf disks. A. 4-weeks old Arabidopsis thaliana accession Col-0 grown under the growth conditions described in Equipment section. B. Cut two leaf disks (7.5 mm diameter) from one plant (one disk per leaf) using a cork borer on paper towels. C. Float two leaf disks (adaxial surface down) on 2 ml of sterilized ultrapure water in one well of a 12-well cell culture plates. D. Drop 100 µl of the water from one well with a pipette onto the sensor of an electrical conductivity meter and measure its conductivity. - Return the sampled water to the well to maintain the water at constant volume during the time course.
- Continue incubating the plate and repeating Steps B8-B9 at additional time points.
Note: The sensor of the electrical conductivity meter should be rinsed with sterilized ultrapure water between one sample and the other.
- Cut two leaf disks (7.5 mm diameter) from one plant (one disk per leaf) using a cork borer on paper towels (Figure 1B). Leaf disks should be cut out before the infiltrated bacterial suspension from Step A7 dries out from the intercellular space of the leaves, which occurs approximately 20 min after infiltration.
Data analysis
- For data analysis, directly use the values of the conductivity value obtained from the sensor of the electrical conductivity meter. We recommend inclusion of uninoculated leaf disks in the experiment as a negative control.
Note: Data analysis is done using Microsoft Excel. - The experiments were performed at least three independent times, each of which had four to six biological replicates. For statistical analysis, Student’s t-test was used for comparison between two genotypes and a one-way analysis of variance (ANOVA) was used for comparison of more than two genotypes. The increased electrolytic conductivity is interpreted as the extent of cell death (Figure 2).
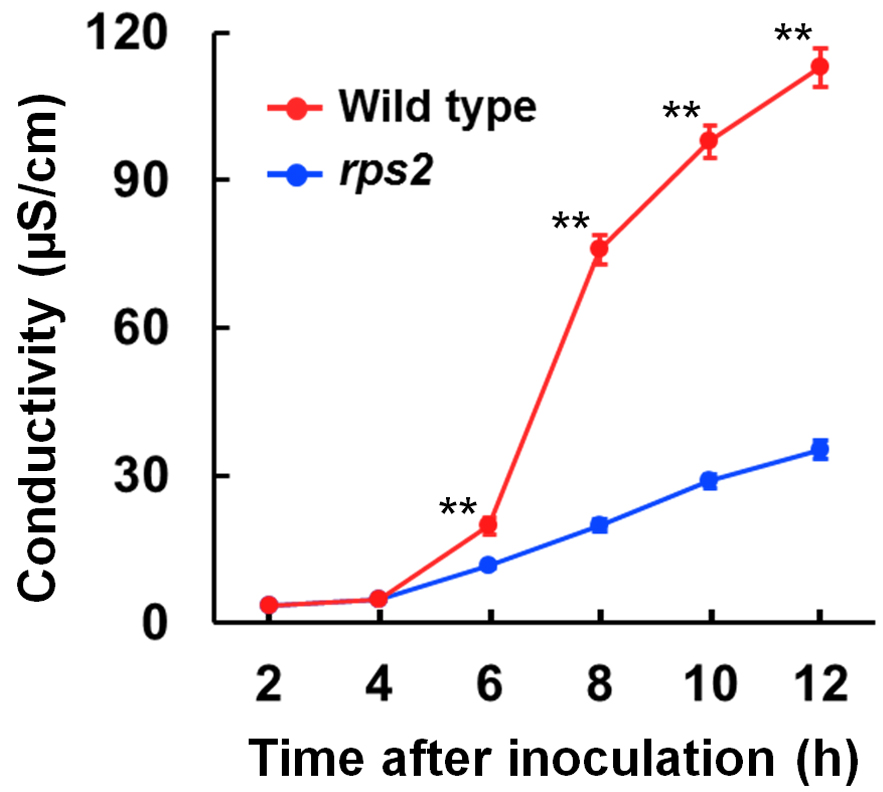
Figure 2. Electrolyte leakage from leaf disks at 2 to 12 h after bacterial inoculation. Representative data of electrolyte leakage from leaf disks. Pto DC3000 avrRpt2 was infiltrated into Arabidopsis leaves of wild-type and rps2 mutant plants. Electrolytic conductivity dramatically increased with wild-type plants after bacterial inoculation, whereas the increase was limited with rps2 mutant plants. Error bars indicate standard errors of three independent experiments, each with four biological replicates. Asterisks indicate significant differences compared with rps2 mutant plants (**, P < 0.01, two-tailed t-tests).
Recipes
- King’s B liquid medium
20 g of Bacto proteose peptone No. 3
15 ml of glycerol
1.5 g of dibasic potassium phosphate (K2HPO4)
15 g of Bacto agar
Dissolve the ingredients in distilled water (1 L), adjust the pH of the solution to 7-6.8 with hydrochloric acid (HCl), and then autoclave it
After autoclaving, add 6 ml of 1 M filter-sterilized magnesium sulfate heptahydrate (MgSO4·7H2O) to the 1-L medium
Note: MgSO4·7H2O is added after autoclaving to avoid precipitation. If storing liquid KB medium, do not add MgSO4 until just before use.
Acknowledgments
We thank Jane Glazebrook for critical reading of the manuscript. This work was funded by grants (grant No. IOS-1121425 and MCB-1518058 to F.K) from the National Science Foundation, U.S.A. Many laboratories used similar protocols in the past (Imanifard et al., 2018; Bach-Pages and Preston, 2018). We have adapted this protocol from (Mackey et al., 2002). The authors have no conflicts of interest or competing interests.
References
- Bach-Pages, M. and Preston, G. M. (2018). Methods to quantify biotic-induced stress in plants. Methods Mol Biol 1734: 241-255.
- Bent, A. F., Kunkel, B. N., Dahlbeck, D., Brown, K. L., Schmidt, R., Giraudat, J., Leung, J. and Staskawicz, B. J. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265(5180): 1856-1860.
- Bethke, G., Thao, A., Xiong, G., Li, B., Soltis, N. E., Hatsugai, N., Hillmer, R. A., Katagiri, F., Kliebenstein, D. J., Pauly, M. and Glazebrook, J. (2016). Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28(2): 537-556.
- Demidchik, V., Straltsova, D., Medvedev, S. S., Pozhvanov, G. A., Sokolik, A. and Yurin, V. (2014). Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J Exp Bot 65(5): 1259-1270.
- Hatsugai, N., Hillmer, R., Yamaoka, S., Hara-Nishimura, I. and Katagiri, F. (2016). The μ subunit of Arabidopsis adaptor Protein-2 is involved in effector-triggered immunity mediated by membrane-localized resistance proteins. Mol Plant Microbe Interact 29: 345-351.
- Hatsugai, N., Igarashi, D., Mase, K., Lu, Y., Tsuda, Y., Chakravarthy, S., Wei, H. L., Foley, J. W., Collmer, A., Glazebrook, J. and Kataqiri, F. (2017). A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J 36: 2758-2769.
- Igarashi, D., Bethke, G., Xu, Y., Tsuda, K., Glazebrook, J. and Katagiri, F. (2013). Pattern-triggered immunity suppresses programmed cell death triggered by fumonisin b1. PLoS One 8(4): e60769.
- Imanifard, Z. Vandelle, E. and Bellin, D. (2018). Measurement of hypersensitive cell death triggered by avirulent bacterial pathogens in Arabidopsis. Methods Mol Biol 1743: 39-50.
- Katagiri, F., Thilmony, R. and He, S. Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 1: e0039.
- Mackey, D., Holt III, B. F., Wiig, A. and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108(6): 743-754.
- Mindrinos, M., Katagiri, F., Yu, G. L. and Ausubel, F. M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78(6): 1089-1099.
- Qi, Y., Tsuda, K., Joe, A., Sato, M., Nguyen le, V., Glazebrook, J., Alfano, J. R., Cohen, J. D. and Katagiri, F. (2010). A putative RNA-binding protein positively regulates salicylic acid-mediated immunity in Arabidopsis. Mol Plant Microbe Interact 23(12): 1573-1583.
- Van Poecke, R. M., Sato, M., Lenarz-Wyatt, L., Weisberg, S. and Katagiri, F. (2007). Natural variation in RPS2-mediated resistance among Arabidopsis accessions: correlation between gene expression profiles and phenotypic responses. Plant Cell 19(12): 4046-4060.
- Whitlow, T. H., Bassuk, N. L., Ranney, T. G. and Reichert, D. L. (1992). An improved method for using electrolyte leakage to assess membrane competence in plant tissues. Plant Physiol 98(1): 198-205.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hatsugai, N. and Katagiri, F. (2018). Quantification of Plant Cell Death by Electrolyte Leakage Assay. Bio-protocol 8(5): e2758. DOI: 10.21769/BioProtoc.2758.
Category
Plant Science > Plant immunity > Host-microbe interactions
Plant Science > Plant immunity > Disease bioassay
Biochemistry > Other compound > Ion
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










