- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vivo Analysis of Cyclic di-GMP Cyclase and Phosphodiesterase Activity in Escherichia coli Using a Vc2 Riboswitch-based Assay
(*contributed equally to this work) Published: Vol 8, Iss 5, Mar 5, 2018 DOI: 10.21769/BioProtoc.2753 Views: 7915
Reviewed by: Dennis NürnbergKanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

High-throughput β-galactosidase and β-glucuronidase Assays Using Fluorogenic Substrates
Joshua P. Ramsay
Jul 20, 2013 16150 Views

Real-Time Analysis of Mitochondrial Electron Transport Chain Function in Toxoplasma gondii Parasites Using a Seahorse XFe96 Extracellular Flux Analyzer
Jenni A. Hayward [...] Giel G. van Dooren
Jan 5, 2022 4166 Views

Analysis of Plasmodium falciparum Mitochondrial Electron Transport Chain Activity Using Seahorse XFe96 Extracellular Flux Assays
SaiShyam Ramesh [...] Alexander G. Maier
Nov 5, 2023 2433 Views
Abstract
Cyclic di-guanosine monophosphate (c-di-GMP) is a ubiquitous second messenger that regulates distinct aspects of bacterial physiology. It is synthesized by diguanylate cyclases (DGCs) and hydrolyzed by phosphodiesterases (PDEs). To date, the activities of DGC and PDE are commonly assessed by phenotypic assays, mass spectrometry analysis of intracellular c-di-GMP concentration, or riboswitch-based fluorescent biosensors. However, some of these methods require cutting-edge equipment, which might not be available in every laboratory. Here, we report a new simple, convenient and cost-effective system to assess the function of DGCs and PDEs in E. coli. This system utilizes the high specificity of a riboswitch to c-di-GMP and its ability to regulate the expression of a downstream β-galactosidase reporter gene in response to c-di-GMP concentrations. In this protocol, we delineate the construction of this system and its use to assess the activity of DGC and PDE enzymes.
Keywords: Cyclic di-guanylate monophosphate (c-di-GMP)Background
Cyclic-di-GMP is an important and ubiquitous second messenger in bacteria, which regulates a variety of processes, such as motility-to-sessility transition, biofilm formation, virulence, and cell cycle progression (Römling et al., 2013). The GG(D/E)EF domain has diguanylate cyclase (DGC) activity and it is responsible for the synthesis of c-di-GMP from two GTPs, which is a two-step reaction with 5’-pppGpG as intermediate and two molecules of pyrophosphate as by-products (Ryjenkov et al., 2005). Phosphodiesterases (PDE) with an EAL or an HD-GYP domain hydrolyze c-di-GMP into linear 5’-pGpG (Schmidt et al., 2005) and GMP (Ryan et al., 2006), respectively.
Several tools have been developed to monitor intracellular cyclic di-nucleotide levels and to identify proteins involved in cyclic di-nucleotide signaling, for example, protein-based fluorescence resonance energy transfer (FRET) biosensor (Christen et al., 2010), riboswitch-based fluorescent biosensor (Kellenberger et al., 2015), and riboswitch-based dual-fluorescence reporter (Zhou et al., 2016). However, these tools monitor altered fluorescence of reporters and require the access to flow cytometry or fluorescence microscopy. Here, we report the development of an alternative assay to monitor the intracellular c-di-GMP concentration, namely by monitoring the alteration in β-galactosidase activity in agar-growing cells. For that, the Vc2 riboswitch (Sudarsan et al., 2008) is fused translationally to lacZY and integrated into the chromosome of E. coli strain TOP10. Vc2 is an ‘off’ riboswitch from Vibrio cholerae and thus down-regulates the expression of β-galactosidase when c-di-GMP is bound (Figure 1). The stable integration into the Tn7 attachment site in the chromosome of E. coli avoids copy number effects and eliminates the need to use an antibiotic resistance marker. Changes in c-di-GMP levels are subsequently translated to the alteration in β-galactosidase expression, which is reflected by the color change of the colony growing on an agar plate containing 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). This assay can be used, for instance, to reveal the function of proteins under physiological condition and to assess the enzymatic activity of proteins that are challenging to be purified and tested in vitro. However, the Vc2-based assay described here is a qualitative assessment of the change in intracellular c-di-GMP concentration. Quantification is not crucial for a screening assay, but would be advantageous in, for example, measuring the activity of enzymes. We have demonstrated that the Vc2-based assay can be exploited to verify the activity of both DGCs and PDEs in vivo (El Mouali et al., 2017).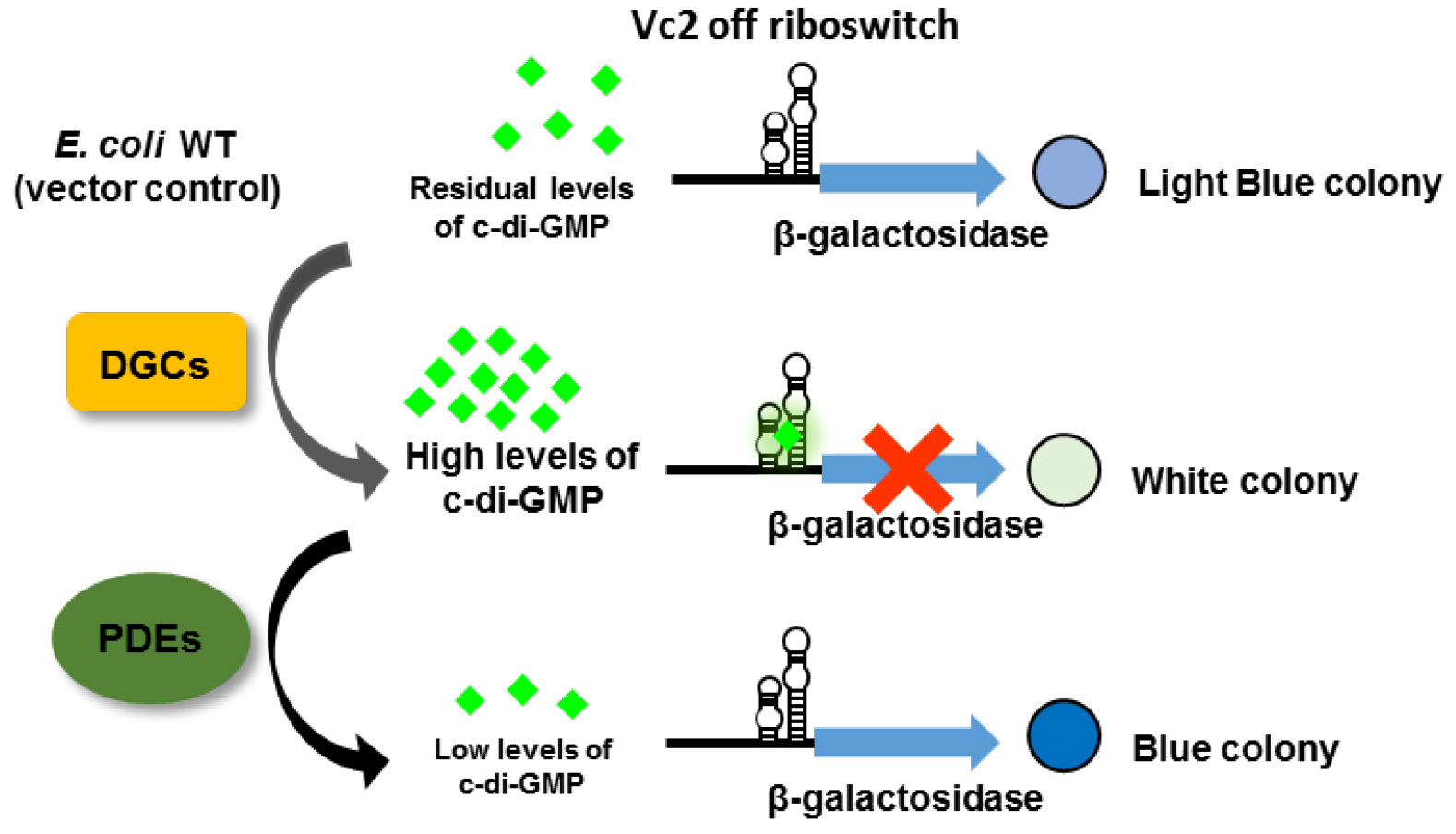
Figure 1. The principle of a riboswitch-based screening system. The Vc2 riboswitch is located upstream of the β-galactosidase open reading frame to control its expression in response to the variation in c-di-GMP concentration. When c-di-GMP is present in high abundance due to the overexpression of DGCs, the expression of β-galactosidase is down-regulated resulting in a white colony on an X-gal containing plate. In contrast, when PDEs are overexpressed, generating low intracellular c-di-GMP concentration, the colony is blue. In E. coli TOP10 wild type cells, there are residual amounts of c-di-GMP, resulting in a light blue colony (adapted from El Mouali et al., 2017).
Materials and Reagents
- PCR tubes
- Petri dish 92 x 16 mm (SARSTEDT, catalog number: 82.1472.001 )
- Syringe filters, 0.2 μm pore size (SARSTEDT, catalog number: 83.1826.001 )
- Microcentrifuge tubes of 1.5 ml (SARSTEDT, catalog number: 72.690.001 )
- Pipette tips (1,000 μl: SARSTEDT, catalog number: 70.762.100 ; 1-200 μl: Corning, catalog number: 4804 ; 0.1-10 μl: Gilson, catalog number: F171103 )
- Spectrophotometer cuvettes (SARSTEDT, catalog number: 67.742 )
- 3-part disposable HSW SOFT-JECT® syringes (5 ml: Henke Sass Wolf, catalog number: 5050.X00V0 ; 10 ml: Henke Sass Wolf, catalog number: 5100.X00V0 )
- Aluminum foil
- Plasmids used in this study:
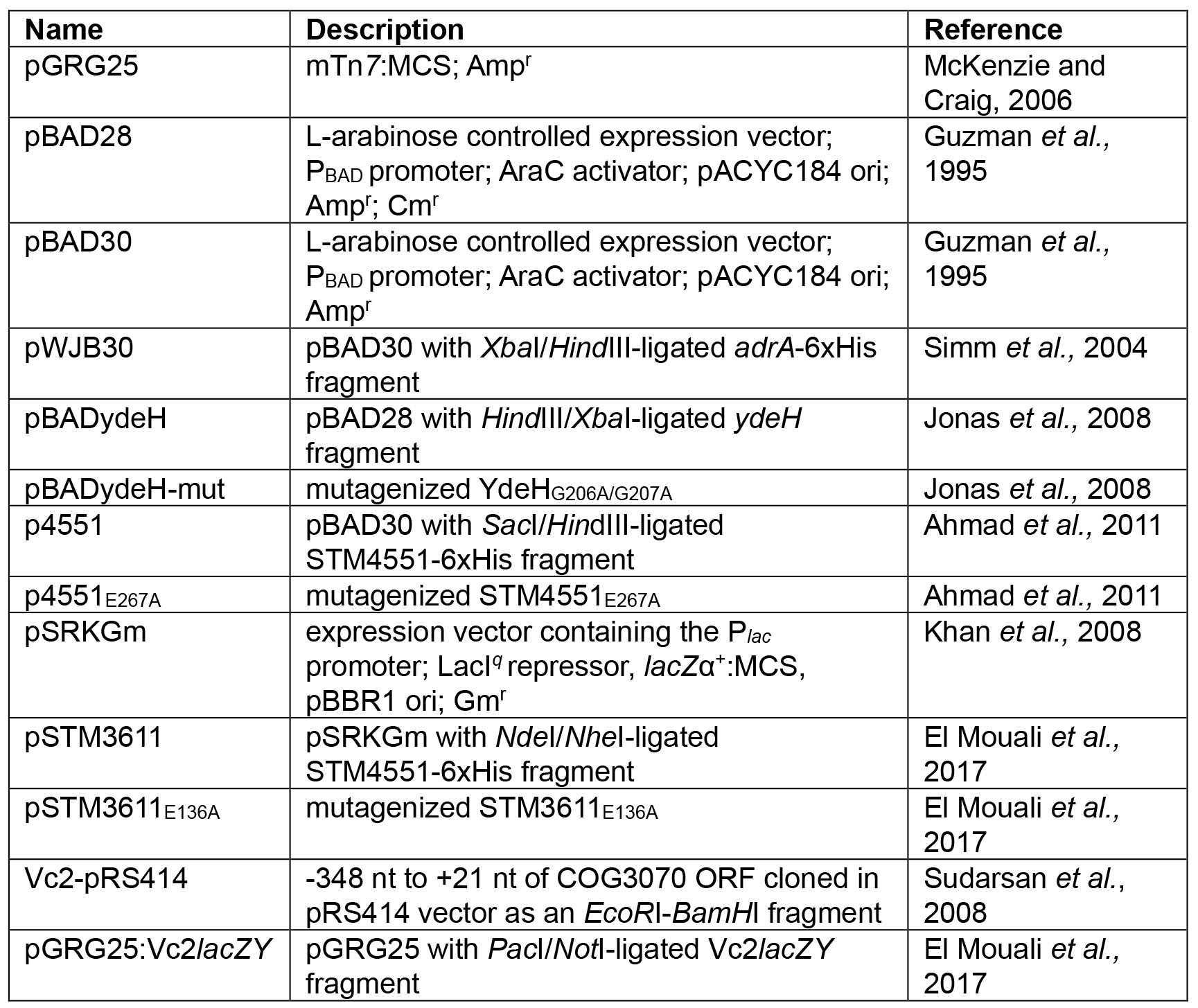
- Calcium chloride competent E. coli TOP10 cells (Hanahan, 1983)
- Primers used in this study (ordered from Sigma-Aldrich):

aThe restriction sites are underlined. - GenElute Plasmid Miniprep Kit (Sigma-Aldrich, catalog number: PLN350 )
- Sterile distilled, deionized water (diH2O)
- Phusion High-fidelity DNA polymerase (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: F530S )
- dNTP mix (10 mM each) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0192 )
- Agarose (Sigma-Aldrich, catalog number: A9539 )
- SmartLadder for DNA: 200-10,000 bp (Eurogentec, catalog number: MW-1700-10 )
- GelRedTM nucleic acid gel stain (Biotium, catalog number: 41003 )
- NotI restriction enzyme (New England Biolabs, catalog number: R0189S )
- PacI restriction enzyme (New England Biolabs, catalog number: R0547S )
- PCR purification kit (QIAGEN, catalog number: 28106 )
- Rapid DNA ligase kit (Roche Diagnostics, catalog number: 10716359001 )
- Taq DNA polymerase with standard Taq buffer (New England Biolabs, catalog number: M0273S )
- Sodium chloride (Sigma-Aldrich, catalog number: S7653 )
- Tryptone (BD, BactoTM, catalog number: 211705 )
- Yeast extract (BD, BactoTM, catalog number: 212750 )
- Agar (BD, DifcoTM, catalog number: 281230 )
- Tris base (Sigma-Aldrich, catalog number: 93350 )
- Glacial acetic acid (Sigma-Aldrich, catalog number: ARK2183 )
- EDTA (Sigma-Aldrich, catalog number: E9884 )
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A9518 )
- Gentamicin sulfate salt (Sigma-Aldrich, catalog number: G3632 )
- L-(+)-arabinose (Sigma-Aldrich, catalog number: A3256 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I6758 )
- Dimethyl sulfoxide (Sigma-Aldrich, catalog number: D8418 )
- 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) (Roche Diagnostic, catalog number: 10703729001 )
- LB (Luria-Bertani) medium (see Recipes)
- LB (Luria-Bertani) agar (see Recipes)
- TAE buffer (see Recipes)
- 100 mg/ml ampicillin stock (see Recipes)
- 30 mg/ml gentamicin stock (see Recipes)
- 100 mM IPTG stock (see Recipes)
- 20 mg/ml X-gal stock (see Recipes)
Equipment
- Erlenmeyer flasks (50 ml/100 ml/250 ml)
- Pipettes [e.g., PIPETMAN® P20 (Gilson, catalog number: F123600 ), P200 (Gilson, catalog number: F123601 ), or P1000 (Gilson, catalog number: F123602 )]
- SureCycler 8800 thermal cycler (Agilent Technologies, model: SureCycler 8800 , catalog number: G8800A)
- HeraeusTM PicoTM 17 Microcentrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM PicoTM 17 , catalog number: 75002410)
- Mini-Sub® cell GT Horizontal Electrophoresis System (Bio-Rad Laboratories, catalog number: 1704406 )
- 15-well comb (Bio-Rad Laboratories, catalog number: 1704464 )
- Sub-Cell GT UV-Transparent Mini-Gel Tray (Bio-Rad Laboratories, catalog number: 1704436 )
- Electrophoresis power supply
- Heraeus® microbiological incubator (Thermo Fisherer Scientific, Thermo ScientificTM, model: Heraeus® microbiological incubator )
- Multitron Standard shaker (INFORS HT)
- Gel DocTM XR+ Gel Documentation System (Bio-Rad Laboratories, catalog number: 1708195 )
- BioPhotometer (Eppendorf, model: 6131 )
- NanoDropTM 2000c Spectrophotometers (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000c , catalog number: ND-2000C)
- Thermomixer compact (Eppendorf, model: 5350 )
- Digital camera
Procedure
Practically, the workflow can be divided into two parts, as shown in Figure 2. The first part is to construct the reporter strain, which takes approximately 12-15 days (Steps 1-3 of the protocol). The second part is to assess the activity of DGC and PDE by the reporter strain (Steps 4-5). This will take 7-9 days.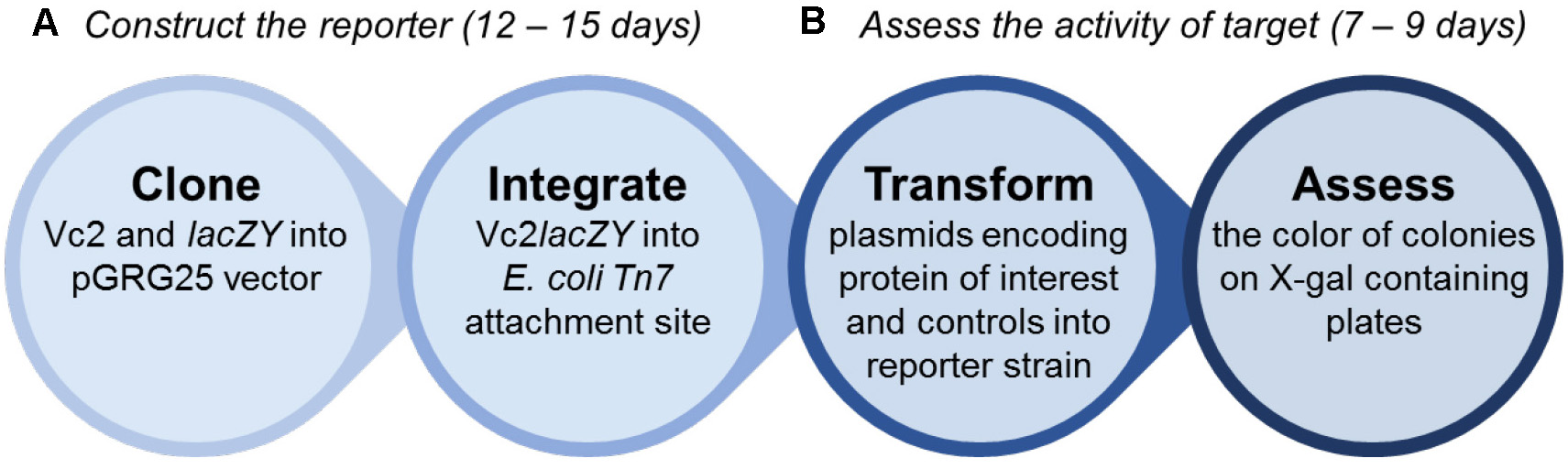
Figure 2. The overview of the workflow. The procedure is divided into two parts: 1) construction of the reporter strain and 2) assessment of c-di-GMP cyclase or phosphodiesterase activity by the reporter. It takes approximately 12-15 days to construct the reporter strain, and another 7-9 days to assess the target.
- Amplify Vc2 and lacZY by PCR:
- The template for the PCR is Vc2-pRS414 (kindly received from Prof. R. R. Breaker, Yale University), in which the fragment containing riboswitch along with its native promoter was cloned in frame to the lacZ reporter gene (Sudarsan et al., 2008).
- The primers used are:
Vc2lacZY_F_PacI: GCGTTAATTAAATAACGCCTATATTTGAAAGCTTG
Vc2lacZY_R_NotI: GATCAGCGGCCGCTTAAGCGACTTCATTCACCTG
The restriction sites PacI and NotI are underlined. - Prepare PCR reaction mix:
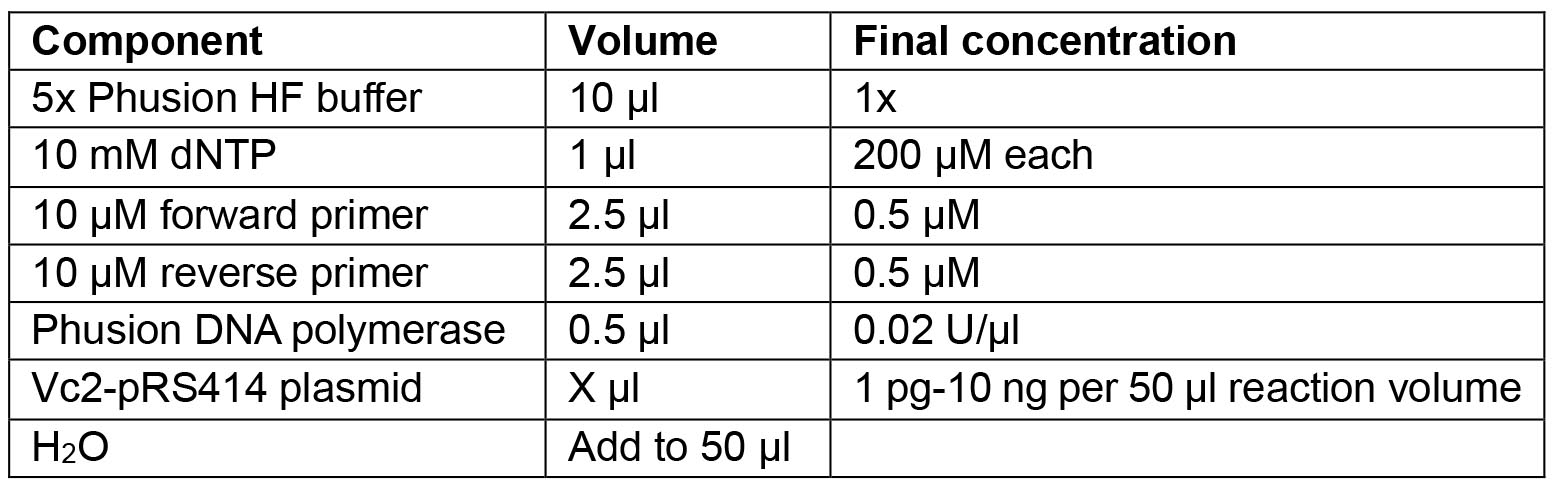
- Thermocycling conditions:
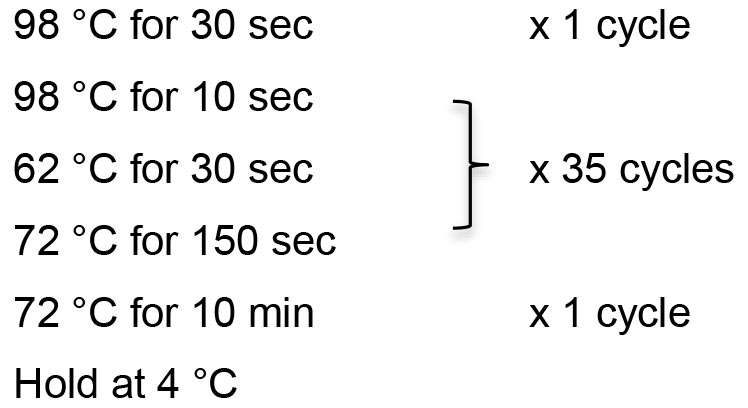
- Check the PCR products by gel electrophoresis using 1% TAE agarose gel. The expected size is 4,733 bp.
- The template for the PCR is Vc2-pRS414 (kindly received from Prof. R. R. Breaker, Yale University), in which the fragment containing riboswitch along with its native promoter was cloned in frame to the lacZ reporter gene (Sudarsan et al., 2008).
- Clone Vc2lacZY into pGRG25 vector:
- Double digest the PCR product Vc2lacZY and the plasmid pGRG25 with PacI and NotI at 37 °C overnight. To reduce star activity, it is advisable to use the fewest units possible to achieve the digestion. After digestion, it is necessary to check the size of products using agarose gel electrophoresis. The enzymes are inactivated at 65 °C for 20 min.
- Remove the restriction enzymes using PCR purification kit. The concentration of digested Vc2lacZY and pGRG25 are measured by NanoDrop.
- Ligate pGRG25 and Vc2lacZY of molar ratio 1:1 at 12 °C overnight using T4 DNA ligase following the manufacturer’s instruction. To verify that the vector is completely cut, ligate the digested vector alone. If the vector is not cut by the two restriction enzymes efficiently, re-circulation will happen and colonies will appear after transformation.
- Transform the ligation products into chemically competent TOP10 cells and select for the recombinant plasmids on LB agar plate containing 100 μg/ml ampicillin. The pGRG25 vector is temperature-sensitive, so the plate should be incubated at 32 °C or even lower temperature.
- Verify the recovered colonies by colony PCR. First, prepare the master mix for Taq DNA polymerase with standard Taq buffer (from New England Biolabs) following manufacturer’s instruction. Then 25 µl aliquots are added to the PCR tubes. Sterile pipette tips are used to pick single colonies and mix with reaction buffer. An initial 5 min denaturation at 95 °C is recommended for colony PCR with Taq DNA polymerase. The expected size is 518 bp. The primers used are:
pGRG25control_F: CACTTATCTGGTTGGTCGACACT
Vc2lacZY_R: CTGCAAGGCGATTAAGTTGG
The details for the colony PCR can be found in the protocol by Azevedo et al. (2017). - Confirm the sequence integrity of the riboswitch in the positive clones by DNA sequencing using the same primers for colony PCR.
- Double digest the PCR product Vc2lacZY and the plasmid pGRG25 with PacI and NotI at 37 °C overnight. To reduce star activity, it is advisable to use the fewest units possible to achieve the digestion. After digestion, it is necessary to check the size of products using agarose gel electrophoresis. The enzymes are inactivated at 65 °C for 20 min.
- Integrate Vc2lacZY into the Tn7 attachment site of E. coli chromosome (Note 1):
- Streak positive colonies on LB agar plate supplemented with 100 μg/ml ampicillin and incubate the cells at 32 °C.
- Inoculate approximately 10 colonies into 3 ml of ampicillin-free LB medium at 32 °C, 200 rpm. Add 0.1% L-arabinose (w/v) to facilitate the incorporation of Vc2lacZY into E. coli Tn7 attachment site. The integration results in a new strain E. coli TOP10 attTn7::Vc2lacZY.
- Dilute the overnight culture by a factor of 108 and plate 50 μl of the diluted culture on LB plate to acquire single colonies. Incubate the plate at 42 °C to block the replication of the temperature-sensitive pGRG25 vector.
- Streak individual colonies on LB plate and repeat the incubation at 42 °C to make sure that the plasmid is lost.
- Confirm the integration of Vc2lacZY into the chromosome by colony PCR using Taq DNA polymerase. The primers used are:
pGRGcontrol_F: GATGCTGGTGGCGAAGCTGT
pGRGcontrol_R: GATGACGGTTTGTCACATGGA
The expected size for positive clone and control is 6,328 bp and 678 bp, respectively.
- Streak positive colonies on LB agar plate supplemented with 100 μg/ml ampicillin and incubate the cells at 32 °C.
- Assess the activity of diguanylate cyclases by Vc2-based screening: take the established DGC AdrA, YdeH and STM4551 as examples
- Prepare calcium chloride competent E. coli TOP10 attTn7::Vc2lacZY cells.
- Transform pBAD vectors (Note 2) harboring DGC (AdrA, YdeH and STM4551) and catalytically inactive mutants (STM4551E267A and YdeH-mut) as well as empty vectors into the chemically competent cells by heat shock at 42 °C for 40 sec. Add 1 ml LB medium free of antibiotics into the microcentrifuge tubes and incubate the tubes at 37 °C, 200 rpm for 1 h to let the cells recover.
- Plate 200 μl cells on LB agar plates containing 100 μg/ml ampicillin and incubate the plates at 37 °C overnight.
- Streak single colonies on ampicillin-containing LB agar plates.
- Pick one colony for each strain and culture the colony in 5 ml of LB medium with 100 μg/ml ampicillin at 37 °C, 200 rpm overnight.
- Measure the OD600 of overnight culture and adjust the OD600 to 0.1.
- Grow cells at 37 °C, 200 rpm. When OD600 reaches 0.6-1.0, spot 3 μl culture on LB plates with the addition of 100 μg/ml ampicillin, 80 μg/ml X-gal, 0.1% (w/v) L-arabinose (Note 3). It is important that cells with DGC, corresponding mutant, and vector control have similar OD600.
- Incubate the plates at 28 °C and monitor the color of colonies up to 72 h. A DGC activity will lead to a white colony indicative of high c-di-GMP levels. Take pictures of the plate using a digital camera.
- Prepare calcium chloride competent E. coli TOP10 attTn7::Vc2lacZY cells.
- Investigate the activity of PDE: take the established PDE STM3611 as an example
- The intracellular concentration of c-di-GMP is low in TOP10 cells, therefore Vc2lacZY-harboring cells have a basal level of β-galactosidase activity resulting in a blue colony, which makes it impossible to distinguish the color further enhanced by phosphodiesterase. The activity of PDE can, however, be reliably investigated by co-expression of a DGC. Therefore, pWJB30 (DGC AdrA cloned into pBAD30) and pSTM3611 (PDE STM3611 cloned into pSRKGm) (Notes 2 and 4) are co-transformed into chemically competent TOP10::Vc2lacZY cells. As controls, empty vectors are co-transformed or in combination with the DGC/PDE. The combinations are listed in Table 1.
Table 1. The combination of plasmids co-transformed into chemically competent TOP10::Vc2lacZY cells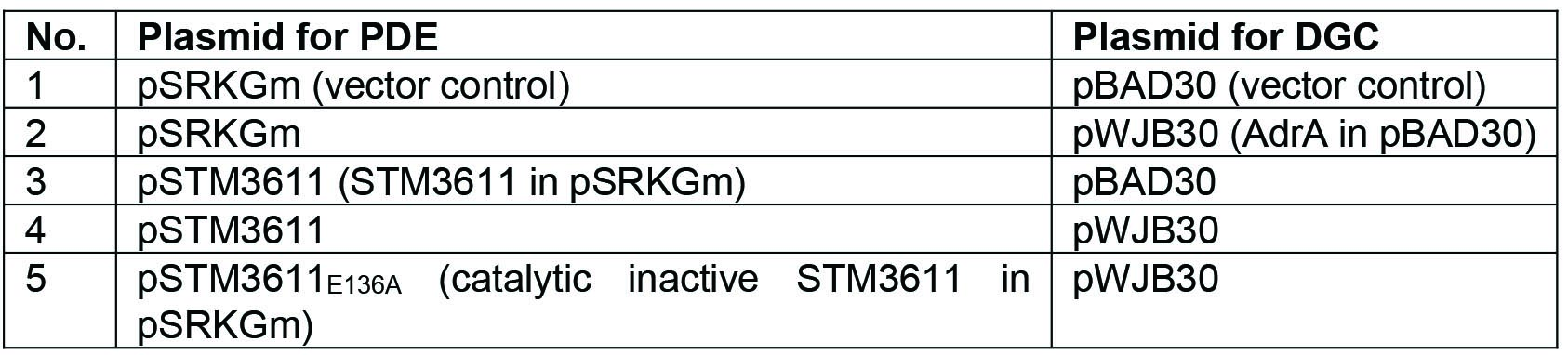
- Plate cells on LB agar plates containing both ampicillin (100 μg/ml) and gentamicin (30 μg/ml). The plates are incubated at 37 °C overnight.
- Streak the positive colonies on antibiotics-containing LB agar plates.
- Inoculate at least three individual colonies per strain in 5 ml of LB medium containing both ampicillin and gentamicin. Grow cells at 37 °C, 200 rpm overnight.
- Measure OD600 of overnight culture and dilute the culture to OD600 of 0.1 with LB medium containing antibiotics. Let the cells grow to exponential phase (OD600 ~0.6). It is critical that all samples have similar OD600 values.
- Spot 3 μl of the cultures on LB agar plates containing ampicillin (100 μg/ml), gentamicin (30 μg/ml), IPTG (0.75 mM), L-arabinose (0.003%, w/v) and X-Gal (80 μg/ml) (Note 3).
- Incubate the plate at 28 °C and record the color of the colonies up to 72 h. While expression of the DGC alone leads to white colonies, co-expression of an active PDE will lead to blue colonies.
- Take pictures of the plate using a digital camera.
- The intracellular concentration of c-di-GMP is low in TOP10 cells, therefore Vc2lacZY-harboring cells have a basal level of β-galactosidase activity resulting in a blue colony, which makes it impossible to distinguish the color further enhanced by phosphodiesterase. The activity of PDE can, however, be reliably investigated by co-expression of a DGC. Therefore, pWJB30 (DGC AdrA cloned into pBAD30) and pSTM3611 (PDE STM3611 cloned into pSRKGm) (Notes 2 and 4) are co-transformed into chemically competent TOP10::Vc2lacZY cells. As controls, empty vectors are co-transformed or in combination with the DGC/PDE. The combinations are listed in Table 1.
Data analysis
- Assessment of the activity of DGC by the Vc2-based assay: after spotted on X-gal containing plates, the color of colonies is monitored periodically. Figure 3 shows the color after 24 h incubation at 28 °C. It is explicit that cells expressing DGCs have whiter color due to the riboswitch inhibition of β-galactosidase expression upon high c-di-GMP levels compared with the corresponding mutant and vector control.
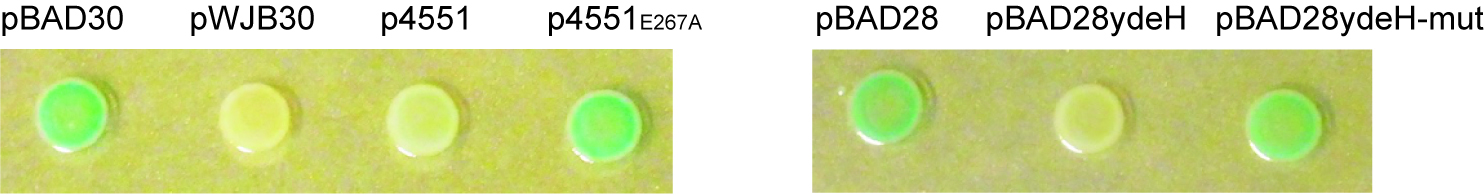
Figure 3. Detection of established diguanylate cyclases by the Vc2-based assay. DGC AdrA, STM4551 and catalytic inactive STM4551E267A were cloned into pBAD30 generating pWJB30, p4551 and p4551E267A, respectively. Another DGC YdeH and its mutant YdeHG206AG207A were cloned into pBAD28 resulting in pBAD28ydeH and pBAD28ydeH-mut. Cells expressing functional DGC (AdrA/STM4551/YdeH) are white, while cells with vector control or catalytically inactive mutant (STM4551E267A/YdeHG206AG207A) are blue. Growth was at 28 °C for 24 h on LB agar plates supplemented with 100 μg/ml ampicillin, 80 μg/ml X-gal and 0.1% arabinose (w/v). - Assessment of the enzymatic activity of a PDE by the Vc2-based assay: Figure 4 shows the color of colonies after incubation at 28 °C for 48 h. Cells expressing the DGC AdrA alone (row 2) or co-expressing AdrA and catalytically inactive PDE STM3611E136A (row 5) are white. However, cells are blue when wild type STM3611 is co-expressed with AdrA (row 4), which illustrates the enzymatic activity of STM3611 and the decrease of the intracellular c-di-GMP concentration upon expression.
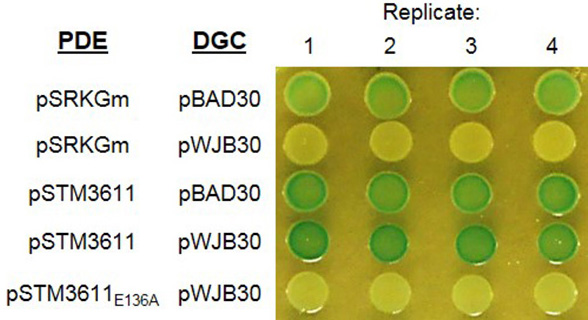
Figure 4. Detection of phosphodiesterase activity of PDE STM3611 by the Vc2-based assay. Phosphodiesterase STM3611 and catalytic inactive STM3611E136A were cloned into pSRKGm generating pSTM3611 and pSTM3611E136A, respectively. Diguanylate cyclase AdrA was cloned into pBAD30 resulting in pWJB30. Cells expressing the DGC AdrA alone have a white color. Upon co-expressing the PDE STM3611 with AdrA, the color turns to blue indicating low c-di-GMP level. However, cells expressing the DGC AdrA with catalytically inactive PDE STM3611E136A are white. Growth was at 28 °C for 48 h on LB agar plate supplemented with 100 μg/ml ampicillin, 30 μg/ml gentamicin, 80 μg/ml X-gal, 0.003% arabinose (wt/vol), and 0.75 mM IPTG (adapted from El Mouali et al., 2017).
Notes
- The protocol for the integration of target genes into the E. coli chromosomal Tn7 attachment site is delineated by G. J. McKenzie and N. L. Craig (McKenzie and Craig, 2006).
- We cloned diguanylate cyclases into pBAD28/pBAD30 vectors and phosphodiesterases into the pSRKGm vector. Other expression vectors can also be used depending on the availability.
- Different L-(+)-arabinose and IPTG concentrations need to be tested to achieve the most discriminating color contrast between diguanylate cyclase, phosphodiesterase and control.
- It is important that diguanylate cyclase and phosphodiesterase are cloned into compatible vectors containing different antibiotic resistance markers and compatible origins of replication.
- The specificity of the Vc2-based riboswitch towards other cyclic di-nucleotides has not been tested in this work.
Recipes
- LB (Luria-Bertani) medium for 400 ml
4 g NaCl
4 g tryptone
2 g yeast extract
Add dH2O to 400 ml
Autoclave at 121 °C for 15 min - LB (Luria-Bertani) agar for 400 ml
4 g NaCl
4 g tryptone
2 g yeast extract
6 g agar
Add dH2O to 400 ml
Autoclave at 121 °C for 15 min
Pour into Petri dishes (add antibiotics, X-gal, L-arabinose, or IPTG if required)
Let cool completely and store at 4 °C - TAE buffer (50x stock solution)
Dissolve 242 g Tris base in water
Add 57.1 ml glacial acetic acid
Add 100 ml of 0.5 M EDTA (pH 8.0)
Add water up to 1 L - 100 mg/ml ampicillin stock
Dissolve 500 mg ampicillin sodium salt in 5 ml deionized water
Filter the solution with 0.2 µm sterile syringe filter
Make 1 ml aliquot of solution into sterile microcentrifuge tubes
Store at -20 °C - 30 mg/ml gentamicin stock
Dissolve 150 mg gentamicin sulfate salt in 5 ml deionized water
Filter the solution with 0.2 µm sterile syringe filter
Make 1 ml aliquot of solution into sterile microcentrifuge tubes
Store at -20 °C - 100 mM IPTG stock
Dissolve 1.191 g IPTG in 5 ml deionized water
Filter the solution with 0.2 µm sterile syringe filter
Make 1 ml aliquot of solution into sterile microcentrifuge tubes
Store at -20 °C - 20 mg/ml X-gal stock
Dissolve 100 mg X-gal in 5 ml DMSO
Wrap the tube with aluminum foil to protect from light
Store at -20 °C
The solution is stable for 6-12 months at -20 °C
Acknowledgments
The authors would like to thank Prof. Dr. Ronald R. Breaker for sharing Vc2-pRS414 riboswitch construct. Prof. Ute Römling conceived this study. Ying Liu developed the assay and assessed DGC activity. Hyunhee Kim developed the assay to assess PDE activity. This work was supported by the Swedish Research Council for Natural Sciences and Engineering (grant 621-2013-4809) and the Karolinska Institutet. This modified protocol is based on a previously published work (El Mouali et al., 2017). The authors declare no conflicts of interest or competing interests.
References
- Ahmad, I., Lamprokostopoulou, A., Le Guyon, S., Streck, E., Barthel, M., Peters, V., Hardt, W. D. and Römling, U. (2011). Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One 6(12): e28351.
- Azevedo, F., Pereira, H. and Johansson, B. (2017). Colony PCR. Methods Mol Biol 1620: 129-139.
- Christen, M., Kulasekara, H. D., Christen, B., Kulasekara, B. R., Hoffman, L. R. and Miller, S. I. (2010). Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328(5983): 1295-1297.
- El Mouali, Y., Kim, H., Ahmad, I., Brauner, A., Liu, Y., Skurnik, M., Galperin, M. Y. and Römling, U. (2017). Stand-alone EAL domain proteins form a distinct subclass of EAL proteins involved in regulation of cell motility and biofilm formation in enterobacteria. J Bacteriol 199(18): pii:e0079-17.
- Guzman, L. M., Belin, D., Carson, M. J. and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177(14): 4121-4130.
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4): 557-580.
- Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Römling, U. and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70(1): 236-257.
- Kellenberger, C. A., Chen, C., Whiteley, A. T., Portnoy, D. A. and Hammond, M. C. (2015). RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP. J Am Chem Soc 137(20): 6432-6435.
- Khan, S. R., Gaines, J., Roop, R. M., 2nd and Farrand, S. K. (2008). Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74(16): 5053-5062.
- McKenzie, G. J. and Craig, N. L. (2006). Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol 6: 39.
- Römling, U., Galperin, M. Y. and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77(1): 1-52.
- Ryan, R. P., Fouhy, Y., Lucey, J. F., Crossman, L. C., Spiro, S., He, Y. W., Zhang, L. H., Heeb, S., Camara, M., Williams, P. and Dow, J. M. (2006). Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103(17): 6712-6717.
- Ryjenkov, D. A., Tarutina, M., Moskvin, O. V. and Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187(5): 1792-1798.
- Schmidt, A. J., Ryjenkov, D. A. and Gomelsky, M. (2005). The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187(14): 4774-4781.
- Simm, R., Morr, M., Kader, A., Nimtz, M. and Römling, U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53(4): 1123-1134.
- Sudarsan, N., Lee, E. R., Weinberg, Z., Moy, R. H., Kim, J. N., Link, K. H. and Breaker, R. R. (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321(5887): 411-413.
- Zhou, H., Zheng, C., Su, J., Chen, B., Fu, Y., Xie, Y., Tang, Q., Chou, S. H. and He, J. (2016). Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter. Sci Rep 6: 20871.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Liu, Y., Kim, H. and Römling, U. (2018). In vivo Analysis of Cyclic di-GMP Cyclase and Phosphodiesterase Activity in Escherichia coli Using a Vc2 Riboswitch-based Assay. Bio-protocol 8(5): e2753. DOI: 10.21769/BioProtoc.2753.
Category
Microbiology > Microbial signaling > Secondary messenger
Microbiology > Microbial cell biology > Cell-based analysis > Reporter assay
Molecular Biology > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











