- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Plate Assay to Determine Caenorhabditis elegans Response to Water Soluble and Volatile Chemicals
Published: Vol 8, Iss 4, Feb 20, 2018 DOI: 10.21769/BioProtoc.2740 Views: 9047
Reviewed by: Gert JansenYan WangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Spatiotemporal Dynamics of Odor Gradient for Small Animals by Gas Chromatography
Akiko Yamazoe-Umemoto [...] Koutarou D. Kimura
Apr 5, 2018 7414 Views

Time-off-pick Assay to Measure Caenorhabditis elegans Motility
Alyssa C. Walker [...] Daniel M. Czyż
Jun 20, 2022 3176 Views

Aerotaxis Assay in Caenorhabditis elegans to Study Behavioral Plasticity
Qiaochu Li [...] Karl Emanuel Busch
Aug 20, 2022 2251 Views
Abstract
The nematode Caenorhabditis elegans is widely used for behavioral studies ranging from simple chemosensation to associative learning and memory. It is vital for such studies to determine optimal concentrations of attractive and aversive chemicals that C. elegans can sense. Here we describe a resource localization assay in which a chemical compound of interest is placed in two compartments of a quadrant plate in order to determine optimal concentrations of the chemical in behavioral studies. Using the assay, we determined the optimal concentration of a water-soluble attractant, KCl, as an unconditioned stimulus for the study of associative learning and memory. In this protocol, we also describe a chemotaxis assay using a square agar plate spotted with an aversive olfactory cue, 1-nonanol, as a conditioned stimulus.
Keywords: AttractantBackground
The nematode Caenorhabditis elegans has extensively been used as a model organism for the study of animal behaviors. C. elegans senses a variety of water-soluble and volatile chemicals that are mainly mediated by amphids, the largest chemosensory organs (Ward, 1973; Dusenbery, 1974; Bargmann and Horvitz, 1991; Bargmann et al., 1993). It is essential for the behavioral study to determine precise concentrations of chemicals that can be sensed by C. elegans. To determine optimal concentrations of water-soluble attractants for C. elegans, Wicks et al. (2000) used a quadrant agar plate for the behavioral assay in which a chemical of interest was mixed with agar in two compartments and this assay has widely been used for many chemicals (e.g., Jansen et al., 2002; Ortiz et al., 2009; Murayama and Maruyama, 2013; Sassa et al., 2013). Chemotaxis assay has also been used to measure the sensitivity of C. elegans to volatile compounds spotted on an agar plate (Bargmann et al., 1993; Troemel et al., 1997). C. elegans is also an excellent model organism for the study of associative learning and memory, in which water-soluble chemicals and volatile chemicals were used as an unconditioned stimulus (US) and a conditioned stimulus (CS) (Amano and Maruyama, 2011; Nishijima and Maruyama, 2017). For effective conditioning of worms, concentrations of CS and US are crucial parameters. The resource localization assay with quadrant agar plates and a chemotaxis assay on square agar plates were successfully used to define optimal concentrations of US and CS for the study of learning and memory. Therefore, these assays could be applied for many other attractive and repulsive chemicals in C. elegans behavioral analysis.
Materials and Reagents
- Latex gloves
- 1.5 ml plastic tubes, sterile (Eppendorf, catalog number: 0030123328 )
- 1.0 ml pipette tips, sterile (Thermo Fisher Scientific, Thermo Scientific, catalog number: H-111-R100NS-Q )
- 0.2 ml pipette tips, sterile (Quality Scientific Plastics, Thermo Fisher Scientific, Thermo Scientific, catalog number: TTW110RS-Q )
- 10 ml Serological pipettes, sterile (As One, catalog number: 2-5237-04 )
- 50 ml Serological pipettes, sterile (As One, catalog number: 2-5237-06 )
- Bottle top 0.2-µm filter units, sterile (Thermo Fisher Scientific, Thermo Scientific, catalog number: 566-0020 )
- Combitips advanced 50 ml, sterile (Eppendorf, catalog number: 0030089480 )
- Petri dishes, sterile (Kord-Valmark, catalog number: 2901 )
- Quadrant Petri dishes, sterile (Kord-Valmark, catalog number: 2913 )
- Square Petri dishes with grids, sterile (Simport, catalog number: D210-16 )
- Wild-type C. elegans strain N2 (available at Caenorhabditis Genetics Center [CGC], https://cbs.umn.edu/cgc/home)
- E. coli OP50 (available at Caenorhabditis Genetics Center [CGC], https://cbs.umn.edu/cgc/home)
- 1-Nonanol (Sigma-Aldrich, catalog number: 131210-100ML )
- Ethanol (99.5%) (Wako Pure Chemical Industries, catalog number: 057-00451 )
- Chloroform (Nacalai Tesque, catalog number: 08401-65 )
- LB medium capsules (MP Biomedical, catalog number: 3002-021 )
- Sodium chloride (NaCl) (Nacalai Tesque, catalog number: 31320-05 )
- Bacto agar (BD, catalog number: 214010 )
- Bacto peptone (BD, catalog number: 211677 )
- Potassium dihydrogen phosphate (KH2PO4) (Nacalai Tesque, catalog number: 28721-55 )
- Di-potassium hydrogen phosphate (K2HPO4) (Nacalai Tesque, catalog number: 28726-05 )
- HEPES (Nacalai Tesque, catalog number: 17514-15 )
- Sodium hydroxide (NaOH) (Wako Pure Chemical Industries, catalog number: 198-13765 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Nacalai Tesque, catalog number: 06730-15 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Nacalai Tesque, catalog number: 21003-75 )
- Potassium chloride (KCl) (Nacalai Tesque, catalog number: 28514-75 )
- D-sorbitol (Sigma-Aldrich, catalog number: S1876-1KG )
- Gelatin (Wako Pure Chemical Industries, catalog number: 073-06295 )
- Cholesterol (Wako Pure Chemical Industries, catalog number: 034-03002 )
- LB broth (see Recipe 1)
- NGM plates (see Recipe 2)
- 1.0 M potassium phosphate (pH 6.0) (see Recipe 3)
- 1.0 M HEPES-NaOH (pH 7.2) (see Recipe 4)
- 1.0 M CaCl2 (see Recipe 5)
- 1.0 M MgSO4 (see Recipe 6)
- Agar for resource localization assay plates (see Recipe 7)
- 2.0% Molten agar (see Recipe 8)
- 0.25% Aqueous gelatin solution (see Recipe 9)
- 5.0 mg/ml cholesterol (see Recipe 10)
- Chemotaxis assay plates (see Recipe 11)
- Doubly deionized water (ddH2O; see Recipe 12)
Equipment
- Safety goggles
- A laboratory coat
- Worm pick
- Dental burner (Phoenix-Dent, model: APT-3 )
- Bunsen burner (EISCO)
- Incubator (SANYO, model: MIR-553 )
- Heating magnetic stirrer (Thermo Fisher Scientific, Thermo Scientific, model: SP131324 )
- Magnetic stirrer bar
- 1.0 L beaker
- Pipet-Aid® XP (Drummond Scientific, model: Pipet-Aid® XP, catalog number: 4-000-101 )
- Multipette M4 (Eppendorf, catalog number: 4982000012 )
- P20 pipetman (Gilson, catalog number: F123600 )
- P100 pipetman (Gilson, catalog number: F123615 )
- P1000 pipetman (Gilson, catalog number: F123602 )
- Kimwipes S-200 (Nippon Paper Crecia, catalog number: 62011 )
- Osmometer (Gonotec, model: Osmomat 030-D )
- Autoclave (Tomy Digital Biology, model: SX-300 )
- Aspirator
- Stereomicroscope (Olympus, model: SZX16 )
- Water purification system (Merck, model: Elix® Essential 10 UV )
- Water purification system (Merck, model: Milli-Q® Synthesis A10® )
Procedure
Part I: Protocol for resource localization assay (the entire procedure is carried out on the bench)
- Preparation of synchronized worms
- Place 10 adult hermaphrodites on an OP50-seeded nematode growth medium (NGM) plate (see Recipe 2), 6 cm in diameter near the Bunsen burner flame, and incubate for 4 h at 20 °C.
- Remove adults from the NGM plates near the Bunsen burner flame, and incubate the plates for 4 days at 20 °C (Note 1).
- Place 10 adult hermaphrodites on an OP50-seeded nematode growth medium (NGM) plate (see Recipe 2), 6 cm in diameter near the Bunsen burner flame, and incubate for 4 h at 20 °C.
- Preparation of resource localization assay plates
- Prepare agar with or without KCl (see Recipe 7).
- Pour agar without KCl, 13 ml each, into two diagonally located compartments in a quadrant dish.
- After solidifying, pour agar with KCl, 13 ml each, to each of two remaining compartments (Figures 1A and 1B) (Note 2).
- Leave the plate without lid at room temperature for 1 h to dry up the agar surface on the bench.
- Connect the compartments by placing a thin layer of 2.0% molten agar (see Recipe 8), using a P1000 pipetman with a blunted tip. Molten agar at 60 °C is poured along plastic separators from the center to the edge of a tilted plate (Figure 1C).
- Leave the plate at room temperature for 5 min before use to solidify the molten agar.
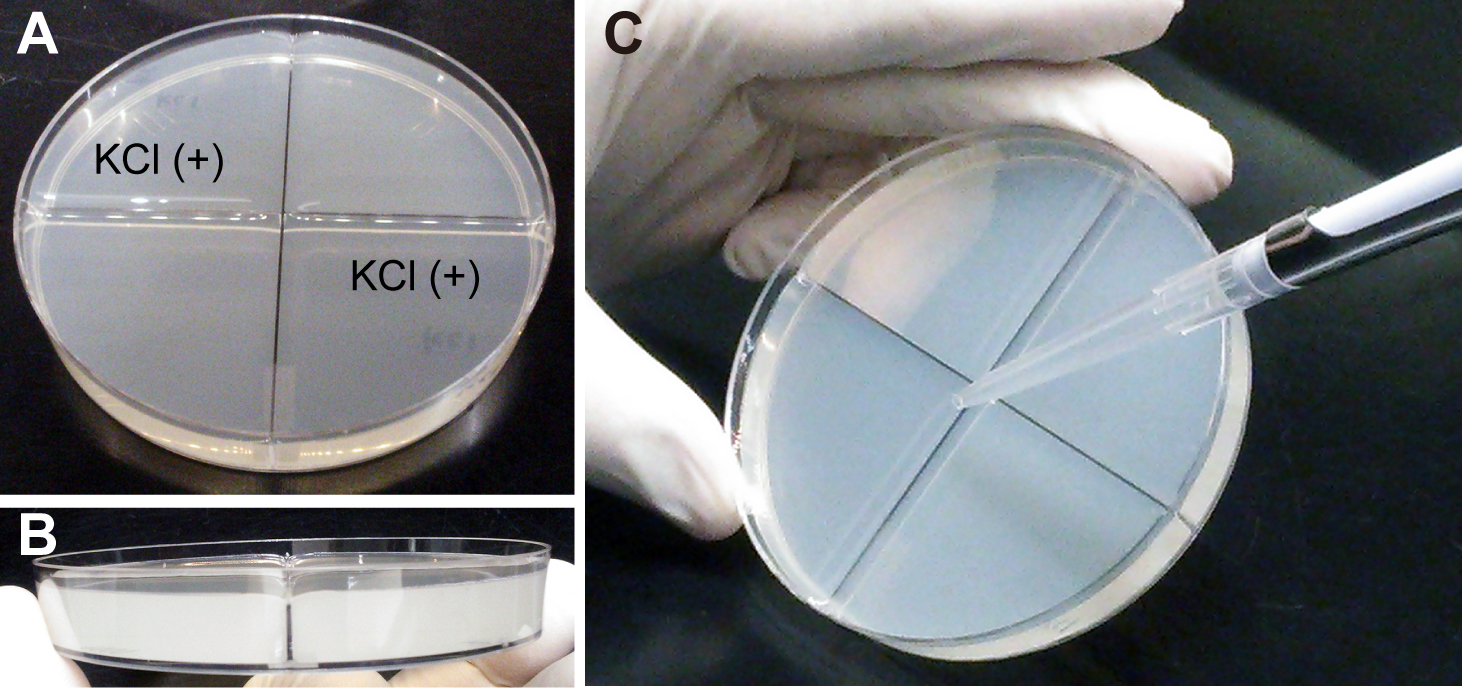
Figure 1. Preparation of resource localization assay plates. A. A quadrant Petri dish poured with agar. KCl (+) indicates agar containing KCl. B. Side-view of the plate. Note that the agar surface is higher than the top of the plastic separator. C. Pouring of 2.0% molten agar on top of the plastic separator from the center to the edge along the separator.
- Prepare agar with or without KCl (see Recipe 7).
- Resource localization assay
- Collect worms from an NGM culture plate, 6 cm in diameter, to a 1.5 ml tube by washing them off with 1.5 ml of 0.25% aqueous gelatin solution (see Recipe 9). After worms have settled at the bottom of the tube (< 1.0 min), remove most of the gelatin solution.
- Wash worms three times with 1.0 ml of 0.25% gelatin solution in the same way as above.
- Resuspend worms at the concentration of 50-100 worms in ~20 µl of gelatin solution.
- Place ~20 µl worm suspension at the center of a quadrant plate, using a P100 Pipetman with a blunted tip.
- Remove as much gelatin solution as possible with a Kimwipes wick.
- Put the lid on the plate, and incubate the plate for 30 min at room temperature.
- Place ~100 µl of chloroform on the lid, and then cover the chloroform with the agar plate to sacrifice worms (Video 1).
Video 1. How to stop worm chemotaxis. At the end of chemotaxis assay, ~100 µl of chloroform was spread on the lid, and then the agar plate was placed upside down on the lid. The same procedure was used for the termination of chemotaxis in Part II. - Count worms in each of the quadrants to calculate a performance index (PI) using the following equation (worms lying against the side of the plate and on the plastic separators are not counted):
PI = [(# of worms in KCl compartments) - (# of worms in reference compartments)]/(# of total worms)
- Collect worms from an NGM culture plate, 6 cm in diameter, to a 1.5 ml tube by washing them off with 1.5 ml of 0.25% aqueous gelatin solution (see Recipe 9). After worms have settled at the bottom of the tube (< 1.0 min), remove most of the gelatin solution.
- Preparation of synchronized worms as described in Part I.
- Chemotaxis assay on a square plate
- Draw lines on a square assay plate (see Recipe 11), 10 x 10 cm, as shown in Figure 2A.
- Prepare worm suspension, which contains ~50 worms per 10 µl, as described above in resource localization assay.
- Place ~10 µl each of the worm suspension at two places as shown in Figure 2A, using a P100 Pipetman with a blunted tip.
- Remove as much gelatin solution as possible with a Kimwipes wick.
- Wait for ~30 sec until worms start moving.
- Spot 3 µl each of 1.0% 1-nonanol diluted with ethanol at two places marked with ‘X’ as shown in Figure 2A, and immediately cover it with a lid.
- Incubate the plate for 10 min at room temperature, and then sacrifice worms with chloroform vapor as described above in resource localization assay.
- Count the number of worms in two areas with and without 1-nonanol to calculate a chemotaxis index (CI) using the following equation (worms lying against the side of the plate and on the lines are not counted):
CI = [(# of worms in odor section) - (# of worms in reference)]/[(# of worms in odor section) + (# of worms in reference)]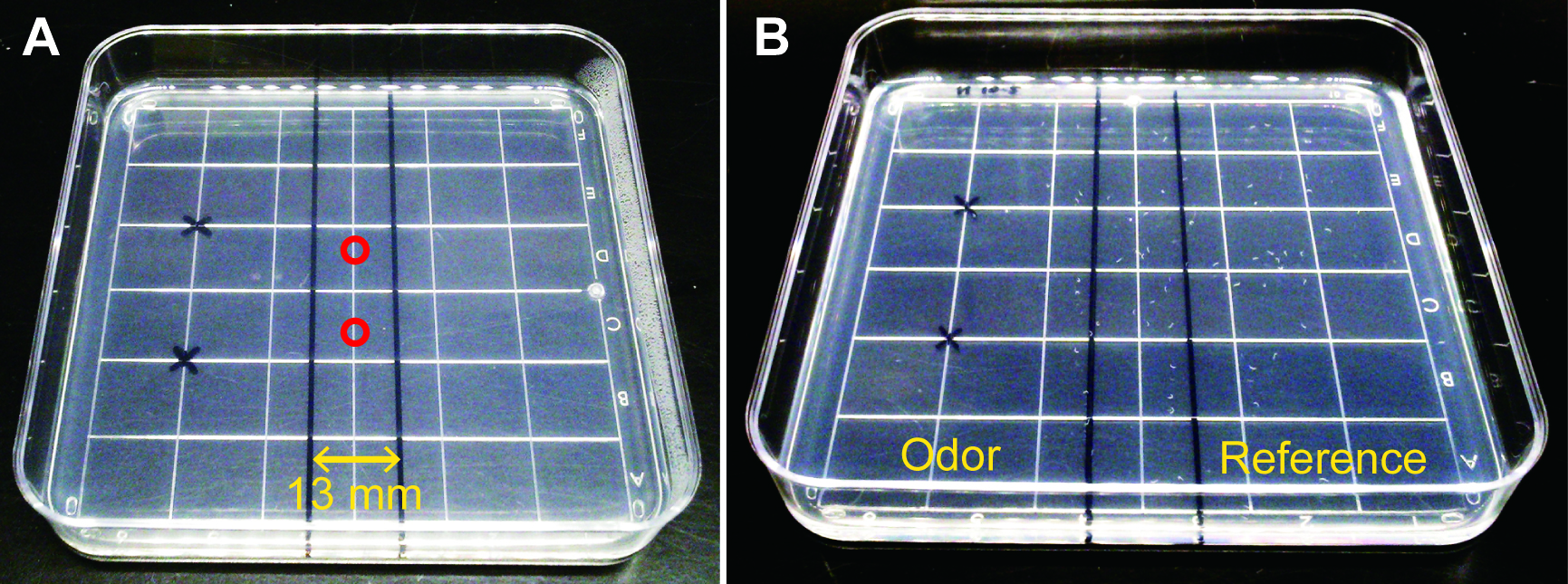
Figure 2. Chemotaxis away from 1-nonanol on a square agar plate. A. A square chemotaxis assay plate marked with red circles showing places where ~10 µl each of worm suspension was spotted, and with ‘X’ indicating positions where 1-nonanol was spotted. B. A typical example of chemotaxis assay after sacrificing worms with chloroform vapor. Note that most of worms (white dots) were repelled from 1-nonanol odor, and mainly located in the reference area on the assay plate.
- Draw lines on a square assay plate (see Recipe 11), 10 x 10 cm, as shown in Figure 2A.
Data analysis
Data analysis should be performed statistically [e.g., using Microsoft® Excel 2011 for Macintosh® with the add-in software Statcel3 (OMS Publ., Saitama, Japan)]. All data should be checked for normality of distribution and homogeneity of variance statistically (e.g., using χ2 goodness of fit test), and should also be evaluated statistically [e.g., using Student’s t-test for comparisons between pairs of groups, or one-way analysis of variance (ANOVA) for multiple comparisons between groups]. If ANOVA results are significant (P < 0.05), the Tukey-Kramer post hoc test or equivalent is used. Results are reported as mean ± the standard error of the mean. The sensitivity of wild-type worms to various concentrations of KCl and 1-nonanol is examined by resource localization assay and chemotaxis assay using square plates, respectively, and is shown in Figure 3.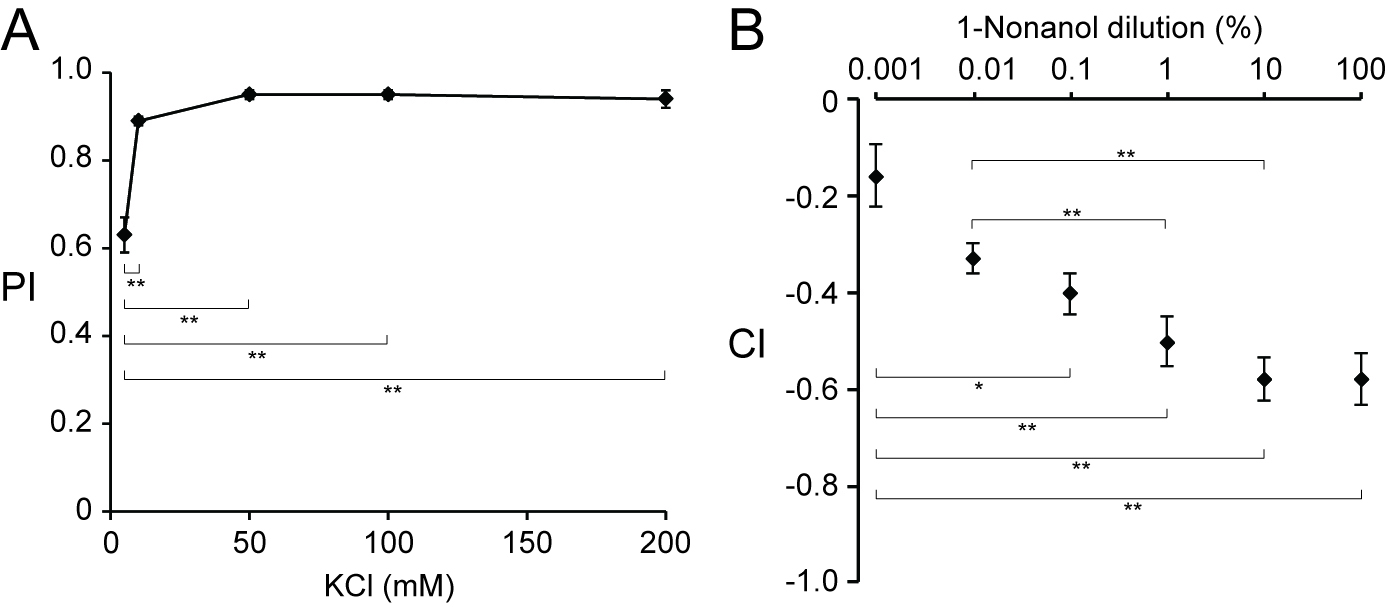
Figure 3. Sensitivity of wild-type C. elegans to various concentrations of KCl and 1-nonanol. A. Performance index (PI) values of wild-type worms, which were measured on resource localization assay plates containing agar with KCl (5-200 mM) or without KCl (n = 6-9 assays). B. Chemotaxis index (CI) values of wild-type worms away from 1-nonanol variously diluted with ethanol, which were assayed using square agar plates (n = 9-15 assays). Asterisks indicate statistically significant (*P < 0.05, **P < 0.01) differences determined by one-way ANOVA, followed by the Tukey-Kramer test for further pair-wise comparisons of all data points. These data are modified from Nishijima and Maruyama (2017).
Notes
- It is easier to remove adult worms from plates by using an aspirator equipped with a 0.2 ml pipette tip than using a worm picker.
- It is convenient to fill quadrant compartments with agar by using a repeating pipette such as Multipette M4. In case that poured agar fused with that in an adjacent compartment, discard the plate since chemicals diffuse out to its adjacent agar.
- 13 ml of agar is required for filling each of four compartments of a quadrant plate, 10 cm in diameter. In this recipe, we describe how to make 500 ml of agar containing 100 mM KCl. Sorbitol was added to reference agar without KCl to give the same osmolality to 100 mM KCl. The osmolality of solutions was measured using an osmometer.
Recipes
- LB broth
- Dissolve 25 capsules of LB medium (see Materials and Reagents) in 1.0 L ddH2O and autoclave at 121 °C for 20 min
- Store at room temperature
- Dissolve 25 capsules of LB medium (see Materials and Reagents) in 1.0 L ddH2O and autoclave at 121 °C for 20 min
- NGM plates
- Dissolve 3 g NaCl, 20 g agar and 2.5 g Bacto peptone in 972 ml doubly deionized water (ddH2O) and autoclave at 121 °C for 20 min
- Let the medium cool to ~60 °C on a heating magnetic stirrer
- Under sterile condition, add 25 ml 1.0 M potassium phosphate (pH 6.0), 1.0 ml 1.0 M CaCl2, 1.0 ml 1.0 M MgSO4 and 1.0 ml 5.0 mg/ml cholesterol in ethanol
- Pour 8 ml of the NGM agar into a Petri dish, 6 cm in diameter, and leave the plate overnight at room temperature
- Prepare an overnight culture of OP50 with LB broth (see Recipe 1), and keep the culture on ice until use
- Spread ~0.1 ml of OP50 culture on the NGM plate under sterile condition, and incubate overnight at room temperature
- Dissolve 3 g NaCl, 20 g agar and 2.5 g Bacto peptone in 972 ml doubly deionized water (ddH2O) and autoclave at 121 °C for 20 min
- 1.0 M potassium phosphate (pH 6.0)
- Dissolve 35.6 g K2HPO4 and 108.3 g KH2PO4 in ddH2O, and adjust the volume to 1.0 L
- After autoclaving at 121 °C for 20 min, store the solution at room temperature
- Dissolve 35.6 g K2HPO4 and 108.3 g KH2PO4 in ddH2O, and adjust the volume to 1.0 L
- 1.0 M HEPES-NaOH (pH 7.2)
- Immediately before use, prepare 1.0 N NaOH by dissolving 12 g NaOH pellets in 300 ml ddH2O
- Dissolve 238.3 g HEPES in ddH2O, and adjust to pH 7.2 with 251 ml 1.0 N NaOH
- Adjust the volume to 1.0 L by adding ddH2O, followed by filtration with a 0.22-µm filter
- Store the solution at room temperature
- Immediately before use, prepare 1.0 N NaOH by dissolving 12 g NaOH pellets in 300 ml ddH2O
- 1.0 M CaCl2
- Dissolve 147 g CaCl2·2H2O in ddH2O, and adjust the volume to 1.0 L
- After autoclaving at 121 °C for 20 min, store at room temperature
- 1.0 M MgSO4
- Dissolve 246.4 g MgSO4·7H2O in ddH2O, and adjust the volume to 1.0 L
- After autoclaving at 121 °C for 20 min, store at room temperature
- Dissolve 246.4 g MgSO4·7H2O in ddH2O, and adjust the volume to 1.0 L
- Agar for resource localization assay plates (Note 3)
- Prepare agar medium containing 100 mM KCl by dissolving 3.7 g KCl and 10 g agar in 494 ml ddH2O by autoclaving at 121 °C for 20 min
- Prepare agar medium without KCl by dissolving 92 ml 1.0 M sorbitol and 10 g agar in 402 ml ddH2O by autoclaving at 121 °C for 20 min
- Let the agar media cool to ~60 °C on a heating magnetic stirrer
- Add 5 ml 1.0 M HEPES-NaOH (pH 7.2), 0.5 ml 1.0 M CaCl2 and 0.5 ml 1.0 M MgSO4 while heating
- Prepare agar medium containing 100 mM KCl by dissolving 3.7 g KCl and 10 g agar in 494 ml ddH2O by autoclaving at 121 °C for 20 min
- 2.0% Molten agar
- Dissolve 2 g agar in 100 ml ddH2O by autoclaving
- Let the solution cool to ~60 °C on a heating magnetic stirrer
- Dissolve 2 g agar in 100 ml ddH2O by autoclaving
- 0.25% Aqueous gelatin solution
- Dissolve 2.5 g gelatin in 1.0 L ddH2O
- After autoclaving at 121 °C for 20 min, store at room temperature
- Dissolve 2.5 g gelatin in 1.0 L ddH2O
- 5.0 mg/ml cholesterol
- Dissolve 250 mg cholesterol in 50 ml of ethanol
- Store at room temperature
- Dissolve 250 mg cholesterol in 50 ml of ethanol
- Chemotaxis assay plates
- Dissolve 15 g agar in 993 ml ddH2O by autoclaving
- Let the solution cool to ~60 °C on a heating magnetic stirrer
- Add 5.0 ml 1.0 M potassium phosphate (pH 6.0), 1.0 ml 1.0 M CaCl2, and 1.0 ml 1.0 M MgSO4
- Pour 14 ml of the solution into a square Petri dish, 10 x 10 cm, and leave the dish on bench overnight
- Dissolve 15 g agar in 993 ml ddH2O by autoclaving
- Doubly deionized water
First, treat tap water with a Millipore Elix 10 UV water purification system, and then with a Millipore Milli-Q Synthesis A10 water purification system
Acknowledgments
A brief version of this protocol was described in Nishijima and Maruyama (2017). This work was partly supported by a JSPS grant (16K07007 to T.M.) and funding to the Information Processing Biology Unit from the Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan. The authors hereby declare no conflict of interest or competing interest.
References
- Amano, H. and Maruyama, I. N. (2011). Aversive olfactory learning and associative long-term memory in Caenorhabditis elegans. Learn Mem 18(10): 654-665.
- Bargmann, C.I., Hartwieg, E. and Horvitz, H.R. (1993). Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74(3): 515-527.
- Bargmann, C. I. and Horvitz, H. R. (1991). Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7(5): 729-742.
- Dusenbery, D. B. (1974). Analysis of chemotaxis in the nematode Caenorhabditis elegans by countercurrent separation. J Exp Zool 188(1): 41-47.
- Jansen, G., Weinkove, D. and Plasterk, R. H. A. (2002). The G-protein gamma subunit gpc-1 of the nematode C.elegans is involved in taste adaptation. EMBO J 21(5): 986-994.
- Murayama, T. and Maruyama, I. N. (2013). Decision making in C. elegans chemotaxis to alkaline pH: Competition between two sensory neurons, ASEL and ASH. Commun Integr Biol 6(6): e26633.
- Nishijima, S. and Maruyama, I. N. (2017). Appetitive Olfactory Learning and Long-Term Associative Memory in Caenorhabditis elegans. Front Behav Neurosci 11: 80.
- Ortiz, C. O., Faumont, S., Takayama, J., Ahmed, H. K., Goldsmith, A. D., Pocock, R., McCormick, K. E., Kunimoto, H., Iino, Y., Lockery, S. and Hobert, O. (2009). Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr Biol 19(12): 996-1004.
- Sassa, T., Murayama, T. and Maruyama, I. N. (2013). Strongly alkaline pH avoidance mediated by ASH sensory neurons in C. elegans. Neurosci Lett 555: 248-252.
- Troemel, E. R., Kimmel, B. E. and Bargmann, C. I. (1997). Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91(2): 161-169.
- Ward, S. (1973). Chemotaxis by the nematode Caenorhabditis elegans: Identification of attractants and analysis of response by use of mutants. Proc Natl Acad Sci USA 70(3): 817-821.
- Wicks, S. R., de Vries, C. J., van Luenen, H. G. A. M. and Plasterk, R. H. A. (2000). CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221(2): 295-307.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Murayama, T. and Maruyama, I. N. (2018). Plate Assay to Determine Caenorhabditis elegans Response to Water Soluble and Volatile Chemicals. Bio-protocol 8(4): e2740. DOI: 10.21769/BioProtoc.2740.
Category
Neuroscience > Behavioral neuroscience > Chemotaxis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










