- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Amyloid Fibril Networks
Published: Vol 8, Iss 4, Feb 20, 2018 DOI: 10.21769/BioProtoc.2733 Views: 9245
Reviewed by: Vivien Jane Coulson-ThomasChristopher J. PoonAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Primary Mouse Choroidal Endothelial Cell Culture
Qiuhua Yang [...] Yuqing Huo
Jun 20, 2025 2149 Views

Spheroid Sheets: A Scalable Platform for Producing Tissue Membrane Constructs
Quang Bach Le [...] Deepak Choudhury
Nov 20, 2025 1536 Views

A Protocol to Induce Brown and Beige Adipocyte Differentiation From Murine and Human Adipose-Derived SVF
Rohit Raj Yadav [...] Narendra Verma
Dec 5, 2025 1642 Views
Abstract
Networks of amyloid nanofibrils fabricated from common globular proteins such as lysozyme and β-lactoglobulin have material properties that mimic the extracellular microenvironment of many cell types. Cells cultured on such amyloid fibril networks show improved attachment, spreading and in the case of mesenchymal stem cells improved differentiation. Here we describe a detailed protocol for fabricating amyloid fibril networks suitable for eukaryotic cell culture applications.
Keywords: Amyloid fibrilsBackground
A wide variety of proteins and peptides can adopt non-native structures and aggregate into amyloid fibrils possessing a common cross β-sheet secondary structure (Nelson et al., 2005). Amyloid fibrils are the pathological hallmarks of a number of neurodegenerative diseases (Chiti and Dobson, 2006), however, there is a growing consensus that mature amyloid fibrils are non-toxic by-products and the toxic species are soluble oligomers, pre-fibrillar aggregates (Kayed et al., 2003), or perhaps the process of aggregation itself (Reynolds et al., 2011). Additionally, a number of functional amyloids with essential physiological features have been discovered in a wide range of organisms (including humans) (Chiti and Dobson, 2006).
Non-toxic synthetic amyloid fibrils can be made from inexpensive, readily available, food grade proteins (Jung et al., 2008; Lara et al., 2011) and have a number of important applications in a wide range of technologies (Dharmadana et al., 2017; Wei et al., 2017). For example 2D and 3D networks of amyloid fibrils have physical and mechanical properties that mimic the local microenvironment of many eukaryotic cells, namely the extracellular matrix (ECM), thus are promising biomimetic materials for cell culture applications (Reynolds et al., 2014 and 2015; Gilbert et al., 2017). Here we describe in detail a protocol to fabricate aqueous suspensions of amyloid fibrils and their subsequent adsorption onto solid supports where they have been shown to control cell adhesion (Reynolds et al., 2014), spreading (Reynolds et al., 2015) and direct the differentiation of Mesenchymal Stem Cells (MSCs) (Gilbert et al., 2017).
Materials and Reagents
- Personal Protective Equipment (PPE): Gloves (latex or nitrile), lab coat and safety glasses
- 15 ml polypropylene centrifuge tubes (Corning, catalog number: 430791 )
- Millex-GP Syringe Filter, 0.22 μm, Polyethersulfone, 33 mm diameter, non-sterile (Merck, catalog number: SLGP033NB )
- Syringe PP/PE without needle, Luer slip tip, 5 ml (Sigma-Aldrich, catalog number: Z116866 )
- Spectra/Por 1 RC Dialysis Membrane Tubing (6-8 kDa MWCO, 25.5 mm diameter) (Fisher Scientific, catalog number: 08-670C)
Manufacturer: Spectrum Medical Industries, catalog number: 132660 . - Dialysis tubing clamps (50 mm) (Sigma-Aldrich, catalog number: Z371092 )
- Pyrex crystallizing dish (used as oil bath) (Capacity 2.5 L) (diameter x height = 190 x 100 mm) (Corning, PYREX®, catalog number: 3140-190 )
- BRAND pipette tips (volume 0.1-20 μl), non-sterile (BRAND, catalog number: 732222 )
- Corning universal fit pipette tips, non-sterile, volume 1-200 μl (Corning, catalog number: 4865 ) and volume 100-1,000 μl (Corning, catalog number: 4867 )
- Muscovite Mica disks, grade V-1 diameter 12.5 mm (ProSciTech, catalog number: G51-12 )
- Double sided tape (for cleaving mica) (Agar Scientific, catalog number: AGG263 )
- Headspace glass vials (20 ml) (Sigma-Aldrich, catalog number: 27306 )
- β-Lactoglobulin from bovine milk ≥ 90% (PAGE), freeze-dried powder (Sigma-Aldrich, catalog number: L3908 )
- Lysozyme from chicken egg white ≥ 90%, freeze-dried powder (Sigma-Aldrich, catalog number: L6876 )
- Hydrochloric acid, ACS Reagent, 37% (Sigma-Aldrich, catalog number: 258148 )
- Sodium hydroxide, BioXtra, ≥ 98% Anhydrous Pellets (Sigma-Aldrich, catalog number: S8045 )
- Silicone oil (Sigma-Aldrich, catalog number: 85409 )
- Hanna pH standard buffer solutions, pH 10 (Sigma-Aldrich, catalog number: Z655155), pH 7 (Sigma-Aldrich, catalog number: Z655139)
Manufacturer: Hanna, catalog numbers: HI 6010 and HI 6007 . - Ricca Chemical Buffer, Reference Standard pH 1.68 (Fisher Scientific, catalog number: 1492-16 )
- 10% lysozyme (or β-Lactoglobulin) solution for purification (see Recipes)
- 2% lysozyme (or β-Lactoglobulin) solution for fibril formation (see Recipes)
Equipment
- BRAND glass beaker with spout 600 ml (BRAND, catalog number: 90648 )
- Hanna bench pH/ISE meter HI4222 (Sigma-Aldrich, catalog number: Z655333EU)
Manufacturer: Hanna Instruments, model: HI-4222 .
Note: This product has been discontinued. - Laboratory centrifuge (Sigma Laborzentrifugen, catalog number: Sigma 3-30KHS )
- 12159 rotor (Sigma Laborzentrifugen, catalog number: 12159 )
- Barnstead E-pure (MilliQ) Water Filtration Device (producing water with a resistivity of ≥ 18.2 MΩ) (Thermo Fisher Scientific, Thermo FisherTM, catalog number: D4631 )
- Labco digital hotplate stirrer (Labtek, catalog number: 400.100.105 )
Note: This product has been discontinued. - Spinbar magnetic stirrer bars PTFE coated polygon 60 x 8 mm (Sigma-Aldrich, catalog number: Z266353 ) and 12.7 x 3 mm (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F37119-0127 )
- Aldrich Clamp Holder (Sigma-Aldrich, catalog number: Z243620 )
- Aldrich Benchclamp 3-prong (Sigma-Aldrich, catalog number: Z556645 )
- Support Stand with Rod (Sigma-Aldrich, catalog number: Z509442 )
- BRAND Ice bucket with lid 4.5 L (BRAND, catalog number: 156100 )
- Gilson PIPETMAN
Classic P20 max volume 20 μl (Gilson, catalog number: F123600 )
Classic P200 max volume 200 μl (Gilson, catalog number: F10005M )
Classic P1000 max volume 1,000 μl (Gilson, catalog number: F123602 ) - Martin Christ Alpha 1-2LDplus Entry Laboratory Freeze Dryer (Martin Christ, model: Alpha 1-2LDplus , catalog number: 101530), equipped with 50 ml single-neck round-bottom flasks (Sigma-Aldrich, catalog number: Z414484 )
- An air or nitrogen sauce (for drying samples, post rinse)
Procedure
- Initial protein purification
To ensure that the final morphology of the amyloid fibrils is as homogenous as possible, commercial grade protein (either hen egg white lysozyme or β-Lactoglobulin) must be further purified to remove trace impurities, such as ash or lipids that can affect fibril morphology. This initial purification is performed by dialysis as originally outlined in Jung et al. (2008). All steps in this protocol should be performed with appropriate Personal Protective Equipment (PPE) (gloves, lab coat and safety glasses).- Calibrate the digital pH meter as per the manufactures instructions using pH 10, 7 and 1.68 calibration standards. Or just pH 7 and 1.68 if three point calibration is not available on the pH meter used.
- Prepare a 10% (weight/weight) solution of protein in fresh MilliQ water (Recipe 1) in 15 ml centrifuge tubes. Once the protein is completely dissolved into the solution, adjust the pH to 4.6 using the digital pH meter, by drop-wise addition of 1 N hydrochloric acid (HCl).
- To remove and precipitate impurities, centrifuge the solution at 20,400 x g (for the 1215-H rotor listed above this is equivalent to 15,000 rpm) for 15 min.
Note: Ensure that the centrifuge is properly balanced before use. Discard the pelleted impurities and add 1 N HCl (dropwise) to the supernatant to adjust to pH 2. - Pass the acidified supernatant through a 0.22 μm filter into a length of the dialysis tubing closed at one end using the dialysis tubing clamp.
Note: Dialysis tubing should be cut to an appropriate length so that it is no more than ~70% full to prevent bursting. - Seal the dialysis tube using a second clamp and immerse the filled tubing in a large beaker filled with an aqueous solution of 0.01 N HCl (dialysing solution), and containing a 60 x 8 mm magnetic stir bar.
Note: The precise volume of the beaker is not important but it should be at least one order of magnitude larger than the volume of dialysate, we typically used beakers with a volume of at least 3 L. - Dialysis should occur at 4 °C, with the dialyzing solution under constant stirring for 5 days. The dialyzing solution should be replaced at least every other day.
Note: Ensure that the fresh dialyzing solution is pre-chilled to 4 °C before switching. - Constant stirring at 4 °C will likely require a refrigerated room that can safely contain a stirrer plate. If this is not available then the dialysis tubing can be kept in a regular refrigerator with no stirring, but it may be necessary to increase the total dialysis time and replace the dialyzing solution more frequently to maintain osmotic pressure.
- After dialysis, the protein solution in the dialysis bag should be readjusted to pH 2 (1 N HCl) and freeze-dried.
Note: Where possible an acid trap should be used with the freeze dryer as prolonged exposure to acidic solutions can lead to corrosion of the equipment. - To freeze-dry the samples, first, to prevent the fibril suspension boiling under reduced vacuum, pre-freeze the fibril suspensions in a regular laboratory freezer. Once frozen switch on the vacuum pump connected to the freeze-dryer and allow it to warm up for 15 min. Turn on the freeze-dryer, select freezing mode and allow the instrument to cool to approximately -55 °C. Connect the round bottom flask containing the frozen fibril suspension to one of the outer ports of the freeze dryer, open the main valve on the vacuum pump and then slowly open the valve on the port with the attached round bottom flask. Switch the instrument to its ‘main drying’ mode, and leave the round bottom flask attached until completely dry (dependent on volume but overnight drying was typically used for 10-20 ml suspensions). At the end of the freeze-drying close the valve to the main pump and release the vacuum in the chamber of the freeze-dryer. If attached to the walls of the round bottom flask, then the freeze-dried fibrils can be gently scraped off with a spatula. The purified, freeze-dried powder can now be stored at -20 °C until it is required for fibril self-assembly.
Note: Never turn off the pump when there is still a vacuum in the chamber as this could result in oil being sucked from the pump.
- Calibrate the digital pH meter as per the manufactures instructions using pH 10, 7 and 1.68 calibration standards. Or just pH 7 and 1.68 if three point calibration is not available on the pH meter used.
- Amyloid fibril self-assembly
All steps in this protocol should be performed with appropriate Personal Protective Equipment (PPE) (gloves, lab coat and safety glasses). As the reaction occurs at 90 °C for 24 h or more, particular care should be taken to ensure the heated oil bath is properly set up and does not pose any danger to yourself or other users of the laboratory.- Dissolve the purified protein in MilliQ water making a 2% solution (weight/weight), using the pH meter to adjust the acidity to pH 2 by dropwise addition of 1 N HCl (Recipe 2). Pass the resulting acidified solution through a 0.22 μm filter into a glass vial, with a heatproof sealable lid (Figure 1).
- Prepare the oil bath by filling an appropriate heat-proof glass beaker (e.g., a crystallization dish) containing a magnetic stirrer bar with clean silicon oil to a level where it will completely cover the protein solution in the glass vial but not totally submerge the vials (Figure 1). Heat the silicon oil in the crystallization dish to 90 °C with gentle stirring (60 rpm, using a digital stirrer hotplate). Stirring is required to ensure homogeneous heating of the oil. At this point, it is important to emphasize that the oil bath should be stable at 90 °C before addition of the protein solution, to ensure constant heating over the entire reaction.
Note: If the hotplate is not equipped with a thermometer and thermostat, then the temperature of the oil bath should be checked and adjusted manually using an external thermometer (deviations in temperature will affect the morphology of the resultant fibrils).
Figure 1. Heated (and stirred) silicon oil bath with a clamped glass vial containing soluble protein/amyloid fibrils - Using a stand and clamp carefully secure the glass vial containing the protein solution in the stirred hot oil bath.
- Fibril formation occurs over time via a progressive denaturing and hydrolysis of the protein and the subsequent self-assembly of the resultant amyloidogenic peptide fragments in a mechanism outlined in Adamcik and Mezzenga (2012). Control over the resultant fibril diameters can be exerted by adjusting the reaction times (Lara et al., 2011; Reynolds et al., 2014). Single protofilaments start to appear after around 5 h; these protofilaments stack together in a modular way (Adamcik et al., 2010; Adamcik and Mezzenga, 2012) to form thicker multi-fibrillar mature fibres over time, before eventually closing up into nanotubes (Lara et al., 2013) or, with the addition of salt [20-50 mM (NaCl) at 2% wt protein], forming hydrogels (Bolisetty et al., 2012). It is worth noting that both the mean diameter and concentration of the fibrils increases over time, therefore it is non-trivial to obtain high concentrations of fibrils composed of only 1 or 2 protofilaments. For cell culture applications, we have found that incubation of lysozyme in the hot oil for between 24-30 h results in fibrils of an optimum concentration and morphology to promote cell growth after adsorption to a 2D substrate (Reynolds et al., 2014; Gilbert et al., 2017). For β-Lactoglobulin fibrils less extensive investigations on the effects of fibril diameter on cell responses have been performed, however fibrils formed after just 5 h incubation have been shown to support MSC culture (Gilbert et al., 2017). Upon removing the protein fibrils from the oil bath, the glass vial should immediately be placed in an ice bath to rapidly quench the reaction and prevent further assembly. Success of the reaction can be qualitatively assessed by observing the color and turbidity of the reaction products. Before heating, the dissolved protein solution should be completely transparent and colorless (Figure 2A), after fibril formation some cloudiness and a yellow hue should be apparent (due to the non-soluble suspension of fibrils) (Figure 2B). For a protocol describing how to quantitatively assess the morphology of the fabricated fibrils by Atomic Force Microscopy (AFM) see Charnley et al. (2018).
- Before use as a cell culture material, suspensions of lysozyme fibrils were dialyzed in order to neutralize the pH. The procedure in Step A4 can be repeated, however now pure MilliQ water should be used as a dialyzing solution and 24 h dialysis with no requirement to change the dialyzing solution is sufficient. This procedure shouldn’t be performed for β-Lactoglobulin fibrils due to concerns about a lack of long-term stability at pH values above its isoelectric point (Jones et al., 2011; Gilbert et al., 2014) instead additional washing steps are employed to remove the acidic solvent after fibril adsorption to the mica discs before incubation with the cells (see Step C2).
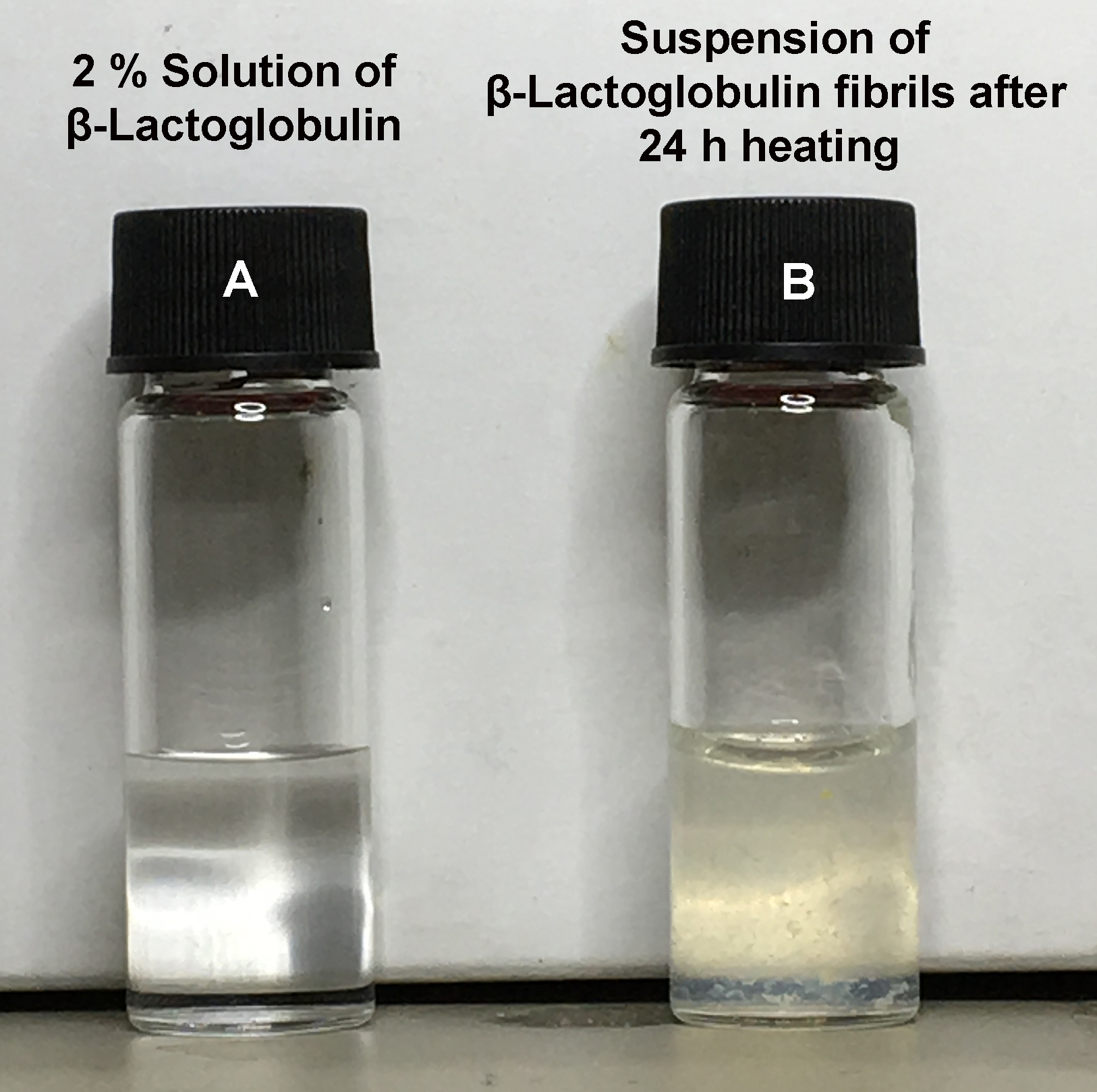
Figure 2. Visual differences between dissolved monomeric protein and amyloid fibril suspension. Photographs of the protein solutions present before heating (A) and the resulting fibril suspension (B) after 24 h of heating. Note the fibril solution has become slightly opaque and has a yellowish hue due to the presence of a suspension of amyloid fibrils.
- Dissolve the purified protein in MilliQ water making a 2% solution (weight/weight), using the pH meter to adjust the acidity to pH 2 by dropwise addition of 1 N HCl (Recipe 2). Pass the resulting acidified solution through a 0.22 μm filter into a glass vial, with a heatproof sealable lid (Figure 1).
- Fabricating Amyloid Fibril Networks
Many solid supports may be suitable for amyloid fibril deposition. However, in our experiments we exclusively use muscovite mica discs (diameter 12 mm). Mica was chosen for a number of reasons. First, at pH 7 lysozyme fibrils carry a net positive charge, and mica carries a net negative charge, thus electrostatic interactions ensure a good coverage of amyloid fibrils on the substrates. Second, mica is composed of many flat sheets held together by weak non-covalent interactions, these sheets can be easily cleaved producing a pristine, dust free surface. Third mica substrates are almost atomically smooth therefore make ideal substrates for characterization by Atomic Force Microscopy (AFM) (Charnley et al. [2018]).
It is worth noting that the first listed benefit of mica (encouraging fibril binding through electrostatic interactions) is only appropriate for lysozyme fibrils, and not β-Lactoglobulin fibrils which have no net charge at neutral pH.- Cleave mica disk. There are many methods of cleaving mica, we prefer to use double-sided tape to stick the disc down to a lab bench and then use a fine tipped pair of tweezers to cleave the top layer from the stuck down disk.
- Deposit 10-20 μl of the 2% (initial monomeric protein concentration) amyloid fibril suspension onto the center of the freshly cleaved mica disc. Incubate at room temperature for 10 min, before gently rinsing the surface of the mica disc with 1 ml of MilliQ water, and gently drying the disc using either a compressed air or nitrogen line. Care should be taken when drying laminated mica discs, to ensure that a homogenous drying is achieved and the dispersed amyloid suspensions are not concentrated at one end of the mica disc during the drying process. Whilst it is not strictly a part of the protocol for fabricating the amyloid fibril networks, it is important to note that immediately before using the fibril networks as substrates for cell culture they should be sterilized to remove any bacterial contamination. In our experiments, we did this by incubating the mica disks with the adsorbed networks in a solution of the antibiotic Pen-Strep (100 ngml-1) for 1 h see Gilbert et al. (2017) for details. Sterilization in an Autoclave was avoided due to concerns that it would damage the fibril network.
- Cleave mica disk. There are many methods of cleaving mica, we prefer to use double-sided tape to stick the disc down to a lab bench and then use a fine tipped pair of tweezers to cleave the top layer from the stuck down disk.
Data analysis
Qualitative analysis of the success of the reaction was performed by simply observing the color and opacity change in the solution before and after incubation in the hot oil bath (Figure 2). A more quantitative analysis of the quality of the fibril coatings produced by atomic force microscopy (AFM) is provided in an associated protocol (Charnley et al. [2018]); and an example AFM image of amyloid nanofibrils adsorbed onto a mica substrate that should be produced from a successful reaction is shown below (Figure 3).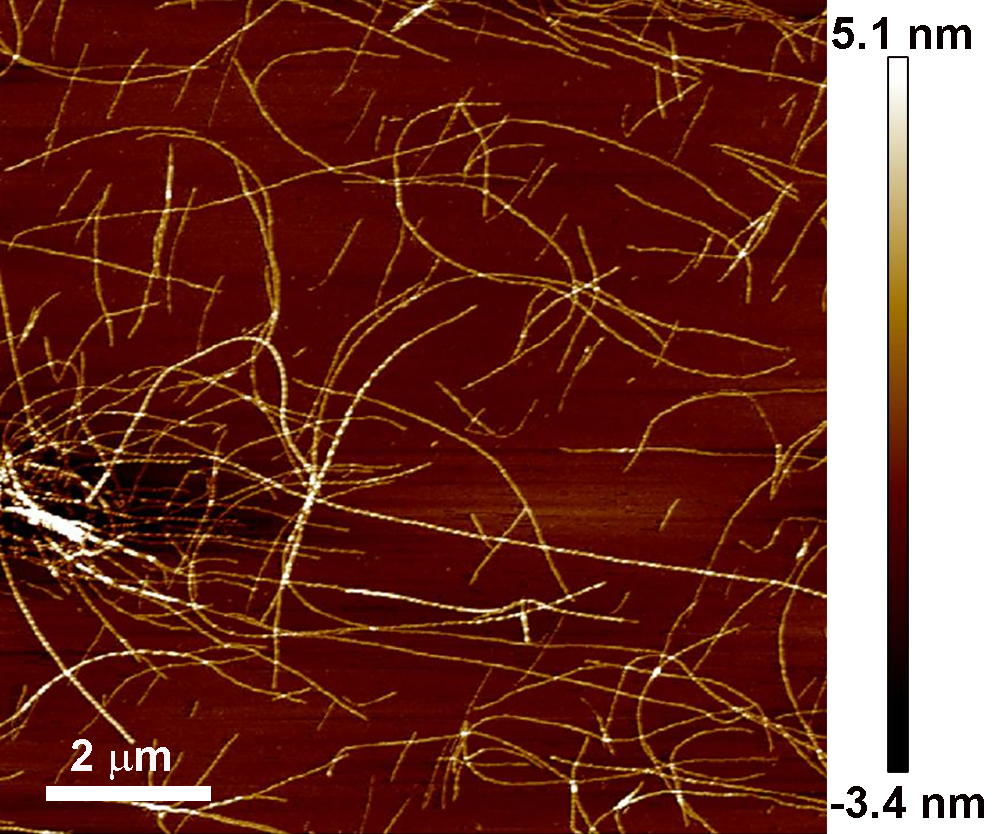
Figure 3. Example of the morphology of amyloid nanofibrils. Produced by the above protocol and imaged by AFM.
Notes
The protocol outlined here is typically very repeatable. However, the final morphology is very sensitive to small variations in temperature throughout the reaction, therefore the hotplate used should maintain a very stable temperature over relatively long periods of time (> 24 h), even small fluctuations in temperature can significantly affect the repeatability of this reaction.
Recipes
- 10% lysozyme (or β-Lactoglobulin) solution for purification
Add 1 part powdered protein to 9 parts MilliQ water (by weight/volume) and adjust to pH 4.6 by the addition of 1 N HCl - 2% lysozyme (or β-Lactoglobulin) solution for fibril formation
Add 1 part purified freeze-dried protein to 49 parts MilliQ water (by weight/volume) and adjust to pH 2 by the addition of 1 N HCl
Acknowledgments
This work was performed in part at the ANFF-Vic node of the Australian National Fabrication Facility, a company established under the National Collaborative Research Infrastructure Strategy to provide nano-and micro-fabrication facilities for Australia’s researchers. JG acknowledges the Australian Government Department of Education and Training for an Endeavour Scholarship and the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1333468. NPR and JG acknowledge the ARC Training Centre for Biodevices at Swinburne University of Technology (IC140100023) for funding (NPR) and for hosting for the duration of his scholarship (JG). MC acknowledges support from the Swiss National Science Foundation (SNSF) (grants PA00P3_142120 and P300P3_154664). JG and OGJ acknowledge further funding support from USDA Hatch Act funds (IND0-1162). The protocols in this work were adapted from a protocol defined in Jung et al. (2008) and Lara et al. (2011). The authors declare no conflict of interest or competing interests.
References
- Adamcik, J., Jung, J. M., Flakowski, J., De Los Rios, P., Dietler, G. and Mezzenga, R. (2010). Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat Nanotechnol 5(6): 423-428.
- Adamcik, J. and Mezzenga, R. (2012). Proteins fibrils from a polymer physics perspective. Macromolecules 45: 1137-1150.
- Bolisetty, S., Harnau, L., Jung, J. M. and Mezzenga, R. (2012). Gelation, phase behavior, and dynamics of beta-lactoglobulin amyloid fibrils at varying concentrations and ionic strengths. Biomacromolecules 13(10): 3241-3252.
- Charnley, M., Gilbert, J., Jones, O. G. and Reynolds, N. P. (2018). Characterisation of amyloid fibril networks by atomic force microscopy. Bio-protocol 8(4): e2732.
- Chiti, F. and Dobson, C. M. (2006). Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: 333-366.
- Dharmadana, D., Reynolds, N. P., Conn, C. E. and Valery, C. (2017). Molecular interactions of amyloid nanofibrils with biological aggregation modifiers: implications for cytotoxicity mechanisms and biomaterial design. Interface Focus 7(4): 20160160.
- Gilbert, J., Campanella, O. and Jones, O. G. (2014). Electrostatic stabilization of beta-lactoglobulin fibrils at increased pH with cationic polymers. Biomacromolecules 15(8): 3119-3127.
- Gilbert, J., Reynolds, N. P., Russell, S. M., Haylock, D., McArthur, S., Charnley, M. and Jones, O. G. (2017). Chitosan-coated amyloid fibrils increase adipogenesis of mesenchymal stem cells. Mater Sci Eng C 79: 363-371.
- Jones, O. G., Handschin, S., Adamcik, J., Harnau, L., Bolisetty, S. and Mezzenga, R. (2011). Complexation of beta-lactoglobulin fibrils and sulfated polysaccharides. Biomacromolecules 12(8): 3056-3065.
- Jung, J. M., Savin, G., Pouzot, M., Schmitt, C. and Mezzenga, R. (2008). Structure of heat-induced beta-lactoglobulin aggregates and their complexes with sodium-dodecyl sulfate. Biomacromolecules 9(9): 2477-2486.
- Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. and Glabe, C. G. (2003). Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300: 486-489.
- Lara, C., Adamcik, J., Jordens, S. and Mezzenga, R. (2011). General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 12(5): 1868-1875.
- Lara, C., Handschin, S. and Mezzenga, R. (2013). Towards lysozyme nanotube and 3D hybrid self-assembly. Nanoscale 5(16): 7197-7201.
- Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R. and Eisenberg, D. (2005). Structure of the cross-[beta] spine of amyloid-like fibrils. Nature 435(7043): 773-778.
- Reynolds, N. P, Charnley, M., Bongiovanni M. N., Hartley, P. G. and Gras, S. L. (2015). Biomimetic topography and chemistry control cell attachment to amyloid fibrils. Biomacromolecules 16(5): 1556-1565.
- Reynolds, N. P., Charnley, M., Mezzenga, R. M. and Hartley, P. G. (2014). Engineered lysozyme amyloid fibril networks support cellular growth and spreading. Biomacromolecules 15(2): 599-608.
- Reynolds, N. P., Soragni, A. Michael, R., Verdes, D., Liverani, E., Handschin S., Riek, R. and Seeger, S. (2011). Mechanism of membrane interaction and disruption by α-synuclein. J Am Chem Soc 133(48): 19366-19375.
- Wei, G., Su, Z., Reynolds, N. P., Arosio, P., Hamley, I. W., Gazit, E. and Mezzenga, R. M. (2017). Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem Soc Rev 46: 4661-4708.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Charnley, M., Gilbert, J., Jones, O. G. and Reynolds, N. P. (2018). Preparation of Amyloid Fibril Networks. Bio-protocol 8(4): e2733. DOI: 10.21769/BioProtoc.2733.
Category
Stem Cell > Adult stem cell > Mesenchymal stem cell
Biochemistry > Protein > Self-assembly
Cell Biology > Cell isolation and culture > Cell growth
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








