- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring Mitochondrial ROS in Mammalian Cells with a Genetically Encoded Protein Sensor
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2705 Views: 10703
Reviewed by: Nicoletta CordaniAlexandros AlexandratosBegona Diaz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 214 Views

Time-Lapse Into Immunofluorescence Imaging Using a Gridded Dish
Nick Lang [...] Andrew D. Stephens
Feb 20, 2026 207 Views

How to Train Custom Cell Segmentation Models Using Cell-APP
Anish J. Virdi and Ajit P. Joglekar
Feb 20, 2026 288 Views
Abstract
Reactive oxygen species (ROS) are not only known for their toxic effects on cells, but they also play an important role as second messengers. As such, they control a variety of cellular functions such as proliferation, metabolism, differentiation and apoptosis. Thus, ROS are involved in the regulation of multiple physiological and pathophysiological processes. It is now apparent that there are transient and local changes in ROS in the cell; in so-called ‘microdomains’ or in specific cellular compartments, which affect signaling events. These ROS hotspots need to be studied in more depth to understand their function and regulation. Therefore, it is necessary to identify and quantify redox signals in single cells with high spatial and temporal resolution. Genetically encoded fluorescence-based protein sensors provide such necessary tools to examine redox-signaling processes. A big advantage of these sensors is the possibility to target them specifically. Mitochondria are essential for energy metabolism and are one of the major sources of ROS in mammalian cells. Therefore, the evaluation of redox potential and ROS production in these organelles is of great interest. Herein, we provide a protocol for the real-time visualization of mitochondrial hydrogen peroxide (H2O2) using the H2O2-specific ratiometric sensor mitoHyPer in adherent mammalian cells.
Keywords: Mitochondrial ROSBackground
ROS are produced as by-products of mitochondrial respiration, through the leakage of electrons from the electron transfer chain. These ROS are considered toxic and cause the oxidation of lipids, proteins, and lead to mitochondrial DNA damage (Ralph et al., 2010; Bogeski and Niemeyer, 2014; Cierlitza et al., 2015; Gibhardt et al., 2016). While mitochondria serve as a hub of metabolism, bioenergetics, and cell death, the emerging role of mitochondrial ROS as second messengers in regulating other cellular functions is also increasingly accepted (Chandel, 2015; Reczek and Chandel, 2015; Shadel and Horvath, 2015; Wilems et al., 2015). To monitor mitochondrial ROS with high spatial and temporal resolution remains challenging due to the short half-life of ROS and the limitation of available probes (Kuznetsov et al., 2011; Norcross et al., 2017). The primary reactive species of mitochondrial origin are superoxide anion, hydroxyl radical, singlet oxygen, and hydrogen peroxide (Gibhardt et al., 2016; Idelchik et al., 2017). Hydrogen peroxide (H2O2) is one of the most stable ROS and is thus an attractive tracking tool for examining the cellular redox state.
During the past decade, several groups designed genetically encoded protein sensors to specifically detect H2O2 (Belousov et al., 2006; Gutscher et al., 2009). The specificity, reversibility, and sensitivity of these protein sensors make them suitable for real-time visualization of H2O2 under a broad range of physiological conditions and stimulations.
The HyPer and roGFP2-Orp1 sensors are advantageous in particular and can be used in various cell systems (Ermakova et al., 2014; Hernandez-Barrera et al., 2013; Bogeski et al., 2016). The HyPer sensor is a combination of a circular permutated yellow fluorescent protein (cpYFP), which is inserted in the regulatory domain of the bacterial H2O2 sensing protein OxyR. The oxidation of cysteine199 found on OxyR initiates conformational changes in HyPer. In a reduced state HyPer has two excitation peaks at 420 nm and 500 nm, and one emission peak at 516 nm. Following oxidation, the peak at 420 nm decreases and the peak at 500 nm increases, thus allowing ratiometric measurement of H2O2. (Bilan and Belousov, 2017). Given that pH fluctuations can also affect the signal from HyPer probes, a mutation at cysteine 199 was introduced to generate a probe named SypHer for monitoring pH, which has the same pH sensitivity but does not react to oxidation (Matlashov et al., 2015; Poburko et al., 2011). The roGFP probe is based on an engineered GFP containing two cysteine residues capable of forming a disulfide bond (Morgan et al., 2011). It has two excitation maxima at 400 and 490 nm with the emission around 510 nm; the ratio of these two excitation maxima depends on the state of the disulfide bond. The development of roGFP probes now provides important alternative tools aimed at detecting H2O2 or the potential of the glutathione redox pair (Gutscher et al., 2008; Kasozi et al., 2013; Habich and Riemer, 2017; Lismont et al., 2017; Müller et al., 2017).
Here we describe a detailed protocol for the real-time imaging and monitoring of mitochondrial H2O2 with the mitoHyPer sensor. The approach can be performed on different cellular systems with a basic understanding of real-time imaging and fluorescence microscopy; the data analysis procedure depends on the software available.
Materials and Reagents
- Round glass coverslips 25 mm No. 1.5 (Kindler/ORSA tec®, Round cover glasses)
- 6-well plates (Corning, Costar®, catalog number: 3516 )
- Falcon tubes (15 ml) (VWR, Corning, catalog number: 62406-200 )
- Serological pipettes (Corning, Costar®, catalog number: 4488 )
- Plasmids
mitoHyPer (Evrogen, catalog number: FP942 )
mitoSypHer (Addgene, catalog number: 48251 ) - Cell growth medium (specific to the cells used in the experiment)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- Fugene® HD (Promega, catalog number: E2312 )
- Opti-MEMTM (Thermo Fisher Scientific, GibcoTM, catalog number: 51985-026 )
- Baysilone paste (VWR, GE Bayer Silicines, catalog number: 291-1210 )
- Accutase (Sigma-Aldrich, catalog number: A6964 ) or Trypsin (Thermo Fisher Scientific, GibcoTM, catalog number: 25300062 )
- 1x DPBS, no calcium, no magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14190-094 )
- 1,4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Hydrogen peroxide solution 30% (w/w) in H2O, contains stabilizer (Sigma-Aldrich, catalog number: H1009 )
- Stimulants and inhibitors (these are experiment-dependent)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Potassium chloride (KCl) (VWR, AnalaR NORMAPUR®, catalog number: 26764.298 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Merck, catalog number: 102382 )
- Magnesium chloride (MgCl2) (Merck, catalog number: 105833025 )
- D(+)-Glucose anhydrous (Merck, catalog number: 108337 )
- EGTA (Sigma-Aldrich, catalog number: E4378 )
- 1 M HEPES (Sigma-Aldrich, catalog number: H7523 )
- Ringer buffer (0.25 mM Ca2+, pH 7.4) (see Recipes)
Equipment
- ZTM Series COULTER COUNTER® Cell (Beckman Coulter, model: 6605699 ) and Particle Counter Z1 (Beckman Coulter or any other counting device)
- Incubator with humidity and gas control for cell culture
- Zeiss Axio Observer.Z1 (Carl Zeiss, model: Axio Observer.Z1 ) setup (Figure 1) (Incubation System S includes Temp Module S, CO2 Module S, O2 Module S, Heating Module S)
- Tweezers (e.g., style Dumont Nr. 7)
- Imaging chamber and ring insert (self-made) and perfusion system (Figure 2)
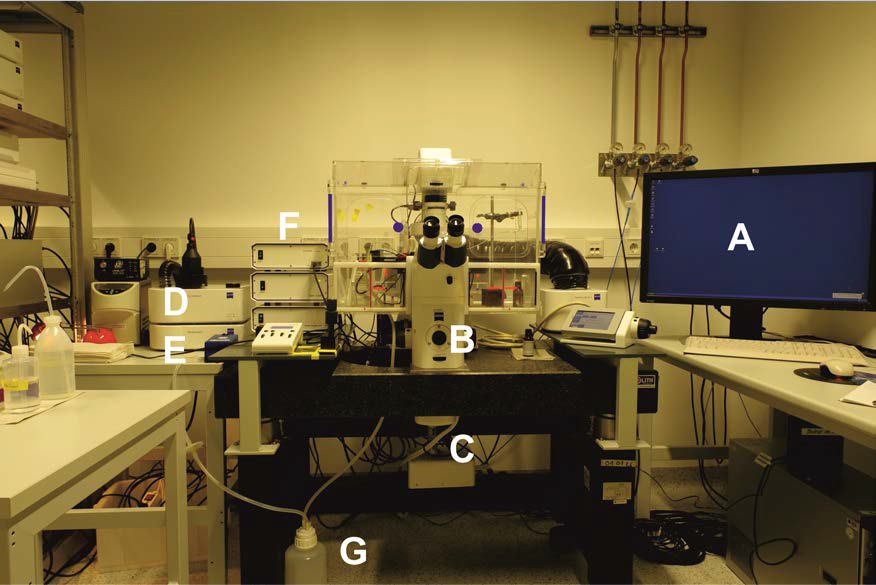
Figure 1. Zeiss Cell Observer.Z1 setup with temperature, CO2 controlling unit, gas chamber and perfusion system. A. Analysis computer; B. Cell Observer.Z1 with 40x oil objective and corresponding filter sets; C. Evolve 512 x 512 EM-CCD camera; D. CO2 supply unit; E. Pecon XL S1 incubator and control modules; F. LED Colibri with corresponding modules. G. Pump and perfusion system.
Notes:- For HyPer measurements, the CFP/YFP filters are essential, but a multiband filter cube with the same property is also a functional option.
- For HyPer experiments, we used the LED light source with the wavelength at 505 nm and 420 nm and corresponding beam splitters.
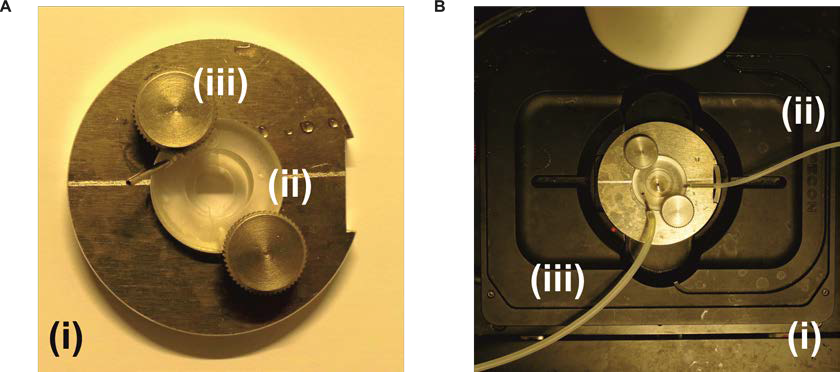
Figure 2. Imaging chamber and mount module with temperature control. A. The self-made imaging chamber (i) with a perfusion chamber plastic insert (ii) that is fixed with knobs (iii). The coverslip with the cells is attached to the lower part of the plastic insert and a small 12 mm coverslip is attached to the upper part of the plastic insert in order to create a small perfusion channel for the measurement. B. The imaging chamber attached to the perfusion system and to the stage of the microscope (i). The perfusion tube (ii) is attached to a syringe in order to add the solutions during the measurement, while the second perfusion tube (iii) is attached to a suction pump system to remove the waste liquid.
- For HyPer measurements, the CFP/YFP filters are essential, but a multiband filter cube with the same property is also a functional option.
Software
- Axiovision 4.6v (Zeiss) with a license for fast acquisition function and measurement analysis or similar
Procedure
- Day 1: Cell culture and seeding
This protocol is exemplary for adherent cells, which can be transfected with reagents such as Fugene® HD. For cell lines which are difficult to transfect, we recommend an alternative transfection method (e.g., nucleofection by electroporation). If stable cells expressing the desired sensors are available, they can also be used for the imaging experiment as described in this protocol.- Culture the cells with their corresponding growth medium until they reach a confluency of around 70%. Remove the growth medium, wash the cells once with 5 ml DPBS and detach the cells by incubating them with 1 ml trypsin or 1 ml accutase (as used for their normal cultivation) at room temperature.
- Suspend the cells in growth medium and dilute 100 µl of the suspension with DPBS to a ratio of 1:100 in a total volume of 10 ml. Determine the concentration of cells in the dilution with a Z1 cell counter or hemocytometer.
- Place autoclaved glass coverslips into a 6-well plate and seed 400,000 cells (in this example HEK293 cells) for each well in 2 ml of growth medium. Place the plate in a humidified cell culture incubator (37 °C, 5% CO2) and incubate overnight.
- Culture the cells with their corresponding growth medium until they reach a confluency of around 70%. Remove the growth medium, wash the cells once with 5 ml DPBS and detach the cells by incubating them with 1 ml trypsin or 1 ml accutase (as used for their normal cultivation) at room temperature.
- Day 2: Transfection
- Remove the Opti-MEM medium and Fugene® HD solutions from the freezer and equilibrate both at room temperature for several minutes.
- Mix 100 µl of Opti-MEM medium with 4 to 10 µl of Fugene® HD solution (according to the manufacturer’s protocol), add the suggested amount of plasmid DNA (1 µg/µl endotoxin-free stock solution) to the mixture (1 µg/well is recommended, but the optimal amount can vary and depends on the cell type and plasmid). Pipette the mix up and down 15 times.
Note: The optimal transfection conditions e.g., cell density, DNA amount, DNA:Fugene® HD ratio might need optimization for the cell line of choice. - Wait for 15 min at room temperature, then add 100 µl of the transfection mixture to each well.
Notes:- If your cell growth medium contains antibiotics, it is advisable to change this before the transfection to growth medium without antibiotics, because they might reduce the transfection efficiency; otherwise, it is not necessary to change the growth medium before the transfection mixture is added.
- Since mitoHyPer and mitoSypHer have the same spectrum features, they should be transfected separately (in different wells).
- If your cell growth medium contains antibiotics, it is advisable to change this before the transfection to growth medium without antibiotics, because they might reduce the transfection efficiency; otherwise, it is not necessary to change the growth medium before the transfection mixture is added.
- The cells are incubated in a humidified cell culture incubator (37 °C, 5% CO2). Change the medium in the transfected wells after 6 h with fresh cell growth medium. Keep the cells in the incubator until ready for imaging (37 °C, 5% CO2), for about 24-48 h.
- Remove the Opti-MEM medium and Fugene® HD solutions from the freezer and equilibrate both at room temperature for several minutes.
- Day 3 or 4: Imaging
Imaging is performed with a Zeiss Cell Observer.Z1 setup with temperature, CO2 controlling unit, gas chamber and perfusion system (Figure 1).- Gently remove a cell-covered coverslip with a pair of delicate tweezers (avoiding the scrapping of cells in the central imaging area of the coverslip). Add Baysilone-paste on the edge of the bottom of the perfusion chamber plastic insert (self-made) and attach it to the coverslip (cells-facing-up). Fix a 12 mm coverslip with Baysilone-paste on the upper part of the plastic insert in order to create a small perfusion channel. Then fix the plastic insert (holding the coverslips) with the knobs and place the assembled chamber into the metal imaging chamber (see Figure 2A).
Note: If simple experiments are performed (e.g., analyzing the resting levels with subsequent addition of saturating H2O2) it might be sufficient to use standard imaging chambers or glass-bottom plates and stimulate the signal changes by addition of the agents carefully with a pipette. However, for more experiments involving, multiple additions or washing-out experiments, a perfusion system is recommended. - Place the imaging chamber on the microscope stage and attach the solution containing perfusion tubes (to avoid air in the system), on opposite sides of the chamber (as shown in Figure 2B). Perfuse gently with 2 ml Ringer solution (see Recipes) to wash away detached cells. Wait for 5 min to reach a CO2 (5 %) and temperature (37 °C) equilibrium before additional handling.
Note: CO2 and temperature are controlled and monitored by the imaging system with the corresponding controlling units. Our perfusion system has on one side a syringe to apply the Ringer solution by hand and on the other side a suction pump to remove the waste. - Using a 40x objective, search for a proper field of view that allows you to assess separate and well attached cells. Set the LED strength in order to get a proper signal, but not too high to avoid photobleaching of the sensor. Optimize the exposure time to obtain good image quality (signal over background) and keep the ratio of exposure time for both channels (420 nm vs. 505 nm) as a constant for all experiments. This part of the procedure will require some time to optimize, based on the cell types used and on the equipment available, since the light source and camera can vary.
- Start the experiment by measuring the resting level of H2O2 in the cells every 1 sec for at least 10 sec, then add stimulating substances through the perfusion system and record until the signal stabilizes (or according to the stimulation protocol). The frame number per minute and total imaging time should be optimized to achieve proper temporal resolution but also to avoid photobleaching.
Note: The stimulating substances leading to the production of ROS from mitochondria vary depending on the scientific question and the cell type. For other scientific questions, only the resting redox level (e.g., the physiological H2O2 concentration under normal conditions) might be of interest. - At the end of each measurement, a single dose of saturating H2O2 (e.g., 1 mM) should be added as a positive control and to determine the maximal intensity of the sensor (this might be needed for calibrating the system). To detect the fluorescence intensity of a fully reduced sensor (which will indicate if the sensor is already oxidized during resting conditions and provide the minimum value for calibration), we advise adding a reducing agent (e.g., 2 mM DTT) at the end of the experiment.
- Perform the same imaging procedure with the mitoSypHer sensor as an imaging control, since the HyPer sensor can be affected by changes in pH.
Note: If the signal (or ratio change) obtained during the experiment with the HyPer sensor is due to oxidation, you will not see any changes in the SypHer signal during the same experimental conditions. If the pH influences your results, you will see also changes in the ratio of the SypHer sensor. Since HyPer is a pH-dependent sensor, this control is mandatory in order to discuss the data regarding redox changes.
- Gently remove a cell-covered coverslip with a pair of delicate tweezers (avoiding the scrapping of cells in the central imaging area of the coverslip). Add Baysilone-paste on the edge of the bottom of the perfusion chamber plastic insert (self-made) and attach it to the coverslip (cells-facing-up). Fix a 12 mm coverslip with Baysilone-paste on the upper part of the plastic insert in order to create a small perfusion channel. Then fix the plastic insert (holding the coverslips) with the knobs and place the assembled chamber into the metal imaging chamber (see Figure 2A).
Data analysis
Analysis with the Axiovision software
- Background correction
The background correction should be performed by subtracting intensity values in a background ROI from a target (cell-based) ROI (Figure 3).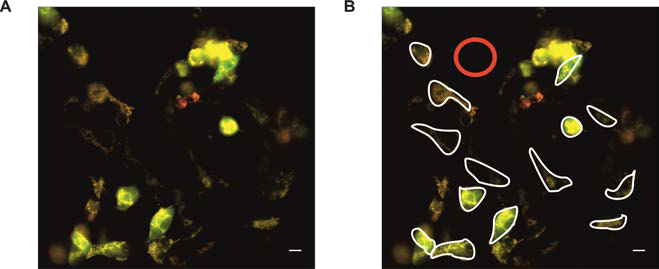
Figure 3. Analysis example of HEK293 cells expressing mitoHyPer (also see Figure 4). A. Merged image (420 nm green, 505 nm red); B. exemplary presentation of analysis. The red circle represents the background ROI in a cell free region, while the borders of some cells are marked with white freehand drawing ROI for analysis. - Ratiometric analysis
The ratio kinetic curve is generated with the equation: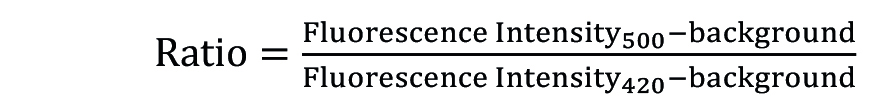
using the Axiovision software.
Note: The data analysis can be performed with different softwares from other suppliers. The basic calculation for the HyPer ratio can also be performed using an open source software such as ImageJ (https://imagej.nih.gov).
The data are usually presented as mean ± SEM (or SD), and tested for significance with two-sided Student’s t-test. For each condition, at least three experiments should be performed with proper replicates. - Below are representative images (A) with a magnification of a single cell (B), a graphed summary (C) and the statistical analysis (D) of what is expected from HEK293 cells following H2O2 and DTT addition (Figure 4).
Note: If the probe is fully oxidized during the measurement and could not respond to saturating H2O2 at the end of the experiment, the result should be excluded from analysis. Since the pH is monitored with the SypHer probe, any experiment with significant fluctuations in pH should be excluded from analysis.
Figure 4. Exemplary ROS measurement of HEK293 cells expressing the mitochondrial H2O2 sensor mitoHyPer. HEK293 cells were transfected with the mitoHyPer sensor using a Fugene® HD-based solution, 48 h prior to imaging. The cells were first titrated with 50 μM and 500 μM H2O2 for oxidation of the probe. Following the washout of H2O2, the cells were titrated with 2 mM DTT for the reduction of the probe. The change in fluorescence intensity ratio is represented as merged images (420 nm green, 505 nm red). As shown in (A), the addition of H2O2 caused oxidation of the probe and increased the signal ratio, while the addition of DTT reduced the signal ratio. The indicated region in (A) is shown magnified in (B). Since mitochondrial H2O2 is generated during a cell’s resting state, the probe can be partially oxidized by the constitutively generated ROS and can be reduced by membrane-permeant reducing agents such as DTT. The time course corresponding to the images in (A) and (B) is shown in (C) and the statistical analysis (mean ± SEM, n = 17) in (D). Scale bars = 10 µm.
Recipes
- Ringer buffer (0.25 mM Ca2+, pH 7.4)
155 mM NaCl
4.5 mM KCl
10 mM glucose
5 mM HEPES
2.75 mM MgCl2
0.25 mM CaCl2
Acknowledgments
This work was supported by the German Research Foundation (DFG) through SFB1190 project 17, SFB1027 project C4 and BO3643/3-2 research grant (all to IB). The authors declare no conflicts of interest or competing financial interests.
References
- Belousov, V. V., Fradkov, A. F., Lukyanov, K. A., Staroverov, D. B., Shakhbazov, K. S., Terskikh, A. V. and Lukyanov, S. (2006). Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 3(4): 281-286.
- Bilan, D. S. and Belousov, V. V. (2017). New tools for redox biology: From imaging to manipulation. Free Radic Biol Med 109: 167-188.
- Bogeski, I. and Niemeyer, B. A. (2014). Redox regulation of ion channels. Antioxid Redox Signal 21(6): 859-862.
- Chandel, N. S. (2015). Evolution of mitochondria as signaling organelles. Cell Metab 22(2): 204-206.
- Cierlitza, M., Chauvistre, H., Bogeski, I., Zhang, X., Hauschild, A., Herlyn, M., Schadendorf, D., Vogt, T. and Roesch, A. (2015). Mitochondrial oxidative stress as a novel therapeutic target to overcome intrinsic drug resistance in melanoma cell subpopulations. Exp Dermatol 24(2): 155-157.
- Gibhardt, C. S., Zimmermann, K. M., Zhang, X., Belousov, V. V. and Bogeski, I. (2016). Imaging calcium and redox signals using genetically encoded fluorescent indicators. Cell Calcium 60(2): 55-64.
- Ermakova, Y. G., Bilan, D. S., Matlashov, M. E., Mishina, N. M., Markvicheva, K. N., Subach, O. M., Subach, F. V., Bogeski, I., Hoth, M., Enikolopov, G. and Belousov, V. V. (2014). Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat Commun 5: 5222.
- Gutscher, M., Pauleau, A. L., Marty, L., Brach, T., Wabnitz, G. H., Samstag, Y., Meyer, A. J. and Dick, T. P. (2008). Real-time imaging of the intracellular glutathione redox potential. Nat Methods 5(6): 553-559.
- Gutscher, M., Sobotta, M. C., Wabnitz, G. H., Ballikaya, S., Meyer, A. J., Samstag, Y. and Dick, T. P. (2009). Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J Biol Chem 284(46): 31532-31540.
- Habich, M. and Riemer, J. (2017). Detection of cysteine redox states in mitochondrial proteins in intact mammalian cells. Methods Mol Biol 1567: 105-138.
- Hernandez-Barrera, A., Quinto, C., Johnson, E.A., Wu, H.M., Cheung, A.Y., and Cardenas, L. (2013). Using hyper as a molecular probe to visualize hydrogen peroxide in living plant cells: a method with virtually unlimited potential in plant biology. Methods Enzymol 527, 275-290.
- Idelchik, M., Begley, U., Begley, T. J. and Melendez, J. A. (2017). Mitochondrial ROS control of cancer. Semin Cancer Biol.
- Kasozi, D., Mohring, F., Rahlfs, S., Meyer, A. J. and Becker, K. (2013). Real-time imaging of the intracellular glutathione redox potential in the malaria parasite Plasmodium falciparum. PLoS Pathog 9(12): e1003782.
- Kuznetsov, A. V., Kehrer, I., Kozlov, A. V., Haller, M., Redl, H., Hermann, M., Grimm, M. and Troppmair, J. (2011). Mitochondrial ROS production under cellular stress: comparison of different detection methods. Anal Bioanal Chem 400(8): 2383-2390.
- Lismont, C., Walton, P. A. and Fransen, M. (2017). Quantitative monitoring of subcellular redox dynamics in living mammalian cells using RoGFP2-based probes. Methods Mol Biol 1595: 151-164.
- Matlashov, M. E., Bogdanova, Y. A., Ermakova, G. V., Mishina, N. M., Ermakova, Y. G., Nikitin, E. S., Balaban, P. M., Okabe, S., Lukyanov, S., Enikolopov, G., Zaraisky, A. G. and Belousov, V. V. (2015). Fluorescent ratiometric pH indicator SypHer2: Applications in neuroscience and regenerative biology. Biochim Biophys Acta 1850(11): 2318-2328.
- Morgan, B., Sobotta, M. C. and Dick, T. P. (2011). Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic Biol Med 51(11): 1943-1951.
- Müller, A., Schneider, J. F., Degrossoli, A., Lupilova, N., Dick, T. P. and Leichert, L. I. (2017). Systematic in vitro assessment of responses of roGFP2-based probes to physiologically relevant oxidant species. Free Radic Biol Med 106: 329-338.
- Norcross, S., Trull, K. J., Snaider, J., Doan, S., Tat, K., Huang, L. and Tantama, M. (2017). Extending roGFP emission via forster-type resonance energy transfer relay enables simultaneous dual compartment ratiometric redox imaging in live cells. ACS Sens 2(11): 1721-1729.
- Poburko, D., Santo-Domingo, J. and Demaurex, N. (2011). Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem 286(13): 11672-11684.
- Ralph, S. J., Rodriguez-Enriquez, S., Neuzil, J., Saavedra, E. and Moreno-Sanchez, R. (2010). The causes of cancer revisited: "mitochondrial malignancy" and ROS-induced oncogenic transformation - why mitochondria are targets for cancer therapy. Mol Aspects Med 31(2): 145-170.
- Reczek, C. R. and Chandel, N. S. (2015). ROS-dependent signal transduction. Curr Opin Cell Biol 33: 8-13.
- Shadel, G. S. and Horvath, T. L. (2015). Mitochondrial ROS signaling in organismal homeostasis. Cell 163(3): 560-569.
- Willems, P. H., Rossignol, R., Dieteren, C. E., Murphy, M. P. and Koopman, W. J. (2015). Redox homeostasis and mitochondrial dynamics. Cell Metab 22(2): 207-218.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, X., Gibhardt, C. S., Cappello, S., Zimmermann, K. M., Vultur, A. and Bogeski, I. (2018). Measuring Mitochondrial ROS in Mammalian Cells with a Genetically Encoded Protein Sensor. Bio-protocol 8(2): e2705. DOI: 10.21769/BioProtoc.2705.
Category
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link