- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Live-cell Imaging of Neisseria meningitidis Microcolony Dispersal Induced by Lactate or Other Molecules
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2695 Views: 5861
Reviewed by: Darrell CockburnTimo LehtiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2895 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2087 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views
Abstract
To efficiently colonize the nasopharyngeal epithelium, the human restricted pathogen Neisseria meningitidis follows a multistep adhesion cascade. First, the bacteria adhere to host cells and aggregate into spherical shaped structures called microcolonies. Several hours later, single bacteria start dispersing from the microcolonies and form a monolayer on top of the host cells. Once in proximity to host cells meningococci can adhere tightly to the epithelial surface or become internalized. This can eventually result in invasion of the mucosal surfaces and gain access to the bloodstream, causing a life-threatening disease. Lactate, a metabolite derived from human epithelial cells, has been previously shown to induce rapid dispersal of N. meningitidis from microcolonies. Here, we describe a host-cell free method based on live-cell imaging to examine the effect of host derived lactate on the timing of N. meningitides microcolony dispersal. Although in this protocol we use lactate, it can be easily modified to test the effects of other molecules.
Keywords: NeisseriaBackground
N. meningitidis is an obligate human pathogen that is responsible for septicemia and/or meningitis. Initial attachment to the nasopharyngeal epithelium and subsequent formation of microcolonies are the first steps in the establishment of infection. In order to cause a disease N. meningitidis must cross the epithelial barrier in the nasopharyngeal mucosa, its natural reservoir, and enter the bloodstream (Stephens, 2009; Trivedi et al., 2011). Dispersal of bacteria from microcolonies plays an important role in progression to an invasive disease as it allows the bacteria to come in close contact with the host epithelium (Pujol et al., 1997 and 1999). Despite its importance, not much is known about the underlying mechanism that governs neisserial detachment from the microcolonies. Recently, we reported that lactate, a common metabolite produced and released from human host cells, is involved in inducing microcolony dispersal (Sigurlásdóttir et al., 2017). Here, we provide a step by step method adapted from Sigurlásdóttir et al. (2017) that can be used to examine the effect of small molecules on N. meningitidis microcolony dispersal in a host-cell free manner by time-lapse imaging. In the described method the focus is to examine microcolony dispersal after addition of lactate, however the effect of other molecules derived from the host or the microbiota can also be tested.
Materials and Reagents
- Glass bottom 24-well cell culture plates (MatTek, catalog number: P24G-1.5-13-F )
- Sterile plastic loops
1 µl plastic loops (SARSTEDT, catalog number: 86.1567.050 )
10 µl plastic loops (SARSTEDT, catalog number: 86.1562.050 )
- Falcon tubes
15 ml Falcon tubes (SARSTEDT, catalog number: 62.554.502 )
50 ml Falcon tubes (SARSTEDT, catalog number: 62.547.254 )
- Pipette tips
0.1-10 µl capacity (SARSTEDT, catalog number: 70.1130.100 )
20-200 µl capacity (SARSTEDT, catalog number: 70.760.502 )
50-1,000 µl capacity (SARSTEDT, catalog number: 70.762.100 )
- Bacteriological Petri plates (SARSTEDT, catalog number: 82.1473 )
- 5-µm pore filter (VWR, catalog number: 514-4106 )
- 5 ml syringe (VWR, catalog number: 613-3940 )
- Serological pipettes
5 ml pipette (SARSTEDT, catalog number: 86.1253.001 )
25 ml pipette (SARSTEDT, catalog number: 86.1685.001 )
- 250 ml vacuum filtration unit, 0.22 μm (SARSTEDT, catalog number: 83.1822.001 )
- Bacterial strain: Neisseria meningitidis serogroup C strain FAM20 (Rahman et al., 1997). FAM20 is a nalidixic acid-resistant mutant of FAM18 (ATCC, catalog number: 700532 )
Note: The bacterial stock is stored in 25% glycerol at -80 °C.
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- GC agar (NEOGEN, catalog number: 7104A )
- D-glucose (Sigma-Aldrich, catalog number: G8270 )
- L-glutamine (Sigma-Aldrich, catalog number: G8540 )
- Ferric(III) nitrate nonahydrate (FeNO3·9H2O) (Sigma-Aldrich, catalog number: F3002 )
Note: This product has been discontinued.
- Cocarboxylase (Sigma-Aldrich, catalog number: C8754 )
- DMEM, no glucose, no glutamine, no phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: A1443001 )
- GlutaMAXTM Supplement (Thermo Fisher Scientific, GibcoTM, catalog number: 35050038 )
- Sodium pyruvate (Thermo Fisher Scientific, catalog number: 11360039 )
- Fetal bovine serum (FBS), heat inactivated (Sigma-Aldrich, catalog number: F9665 )
- Sodium L-lactate (Sigma-Aldrich, catalog number: L7022 )
- Sodium D-lactate (Sigma-Aldrich, catalog number: 71716 )
- GC agar plates (see Recipes)
- 0.2% Cocarboxylase solution (see Recipes)
- Kellogg’s supplement (see Recipes)
- 2.5 mM glucose (see Recipes)
- DMEM (see Recipes)
- DMEM/1%FBS (see Recipes)
- Lactate solutions (see Recipes)
Equipment
- Class II biosafety cabinet (e.g., Esco Micro, model: Airstream® Class II Biological Safety Cabinet )
- Humidified 5% CO2 incubator at 37 °C (e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i )
- Water bath set to 37 °C (e.g., Grant Instruments, model: SAP12 )
- Spectrophotometer (e.g., Bio-Rad Laboratories, model: SmartSpec Plus )
- Inverted live-cell observer, connected to an incubator with a controlled temperature (temperature module) and CO2 module (e.g., Carl Zeiss, model: Axio Observer Z1 )
- Pipette boy (e.g., Fisher Scientific, model: FisherbrandTM Electric Pipet Controller )
- Pipettes
0.5-10 µl capacity (e.g., Eppendorf, catalog number: 4924000029 )
10-100 µl capacity (e.g., Eppendorf, catalog number: 4924000053 )
100-1,000 µl capacity (e.g., Eppendorf, catalog number: 4924000088 )
- 1 L bottle
- 500 ml flask
Software
- AxioVision (Release: 4.8.2 SP3)
- Fiji (http://fiji.sc/; version: ImageJ 2.0.0-rc-44/1.50e)
- Microsoft Excel
Procedure
Caution: N. meningitidis is classified as a class II pathogen. Therefore, all work with bacterial solutions must be done in a Class II biosafety cabinet and according to a national regulation of biosafety. In addition, all bacterial waste should be discarded as Infectious/Biohazardous.
Note: Before performing the experiment the OD600 value corresponding to viable count 108 cfu/ml has to be determined. For the bacterial stock used in this experiment, the OD600: 0.36 is 108 cfu/ml. This can be done by performing serial dilutions of cultures containing bacteria that have been grown on a GC plate for 16-18 h. The dilutions should then be plated on GC plates and incubated overnight at 37 °C with 5% CO2 atmosphere.
Day 1
Preparation of overnight bacterial cultures. Work in a Class II biosafety cabinet.
- N. meningitidis is streaked from a glycerol stock on a GC agar plate (see Recipes) supplemented with 10% Kellogg’s supplement (see Recipes) and incubated overnight for 16-18 h at 37 °C in an incubator with a 5% CO2 environment.
Day 2
- Induction assay: Preparation and incubation of bacterial liquid cultures. Work in a Class II biosafety cabinet.
- The Axio Observer microscope should be turned on and pre-warmed to 37 °C at least 30 min prior to your experiment. The CO2 should be set to 5% before inserting the multiwell plate to the microscope.
- Prepare 30 ml pre-warmed DMEM (see Recipes) containing 1% FBS (DMEM/1% FBS, see Recipes). Fill 900 µl of the medium to the wells of 24-well glass bottom plate (MaTek) that will be used in the experiment (Figure 1). The multiwell plate should be warmed in a 37 °C incubator until needed in Step 1f.
Note: Cultivation of N. meningitidis should always be performed with pre-warmed medium. In addition, the bacterial cultures should never be vortexed prior to experiments as this might induce shedding of bacterial pili, involved in microcolony formation.
- Half fill a 10 µl loop with bacteria from the overnight culture and resuspend in 2 ml of DMEM/1% FBS. Mix the bacterial solution 4 times with a 1 ml pipette. Wait for 3 min and let the largest bacterial aggregates settle down to the bottom of the tube.
- Transfer 1 ml of the uppermost bacterial solution, leaving the large aggregates behind, to a 50 ml Falcon tube containing 5 ml DMEM/1% FBS. Use a syringe to take up the bacterial solution and filter through a 5 µm-pore filter to a 15 ml Falcon tube.
Note: In order to prevent variations in microcolony size that can occur due to differences in bacterial re-suspension, we filter the bacterial solution through 5 µm filter to break all pre-existing aggregates (Eriksson et al., 2012; Engman et al., 2016; Sigurlásdóttir et al., 2017).
- Use a spectrophotometer to measure the absorbance of the bacteria solution. Adjust the absorbance to OD600 = 0.36 (equivalent to 108 cfu/ml) by diluting with DMEM/1%FBS.
- Take the pre-warmed glass bottom plate from the incubator. Add 100 µl of bacterial solution to the wells containing 900 µl of DMEM/1%FBS (Figure 1). Mix 4 times with a 1 ml pipette. Start timing your 3 h incubation exactly when bacterial cultures are added to the multi-well plate. Write down the exact time when the cultures are added to the wells. Keep the 15 ml Falcon tube containing the bacterial solution for viable count (see Step 2a).
- Immediately transfer the multi-well plate to the pre-warmed microscope. Incubate for 3 h at 37 °C and 5% CO2 to allow bacteria to form microcolonies.
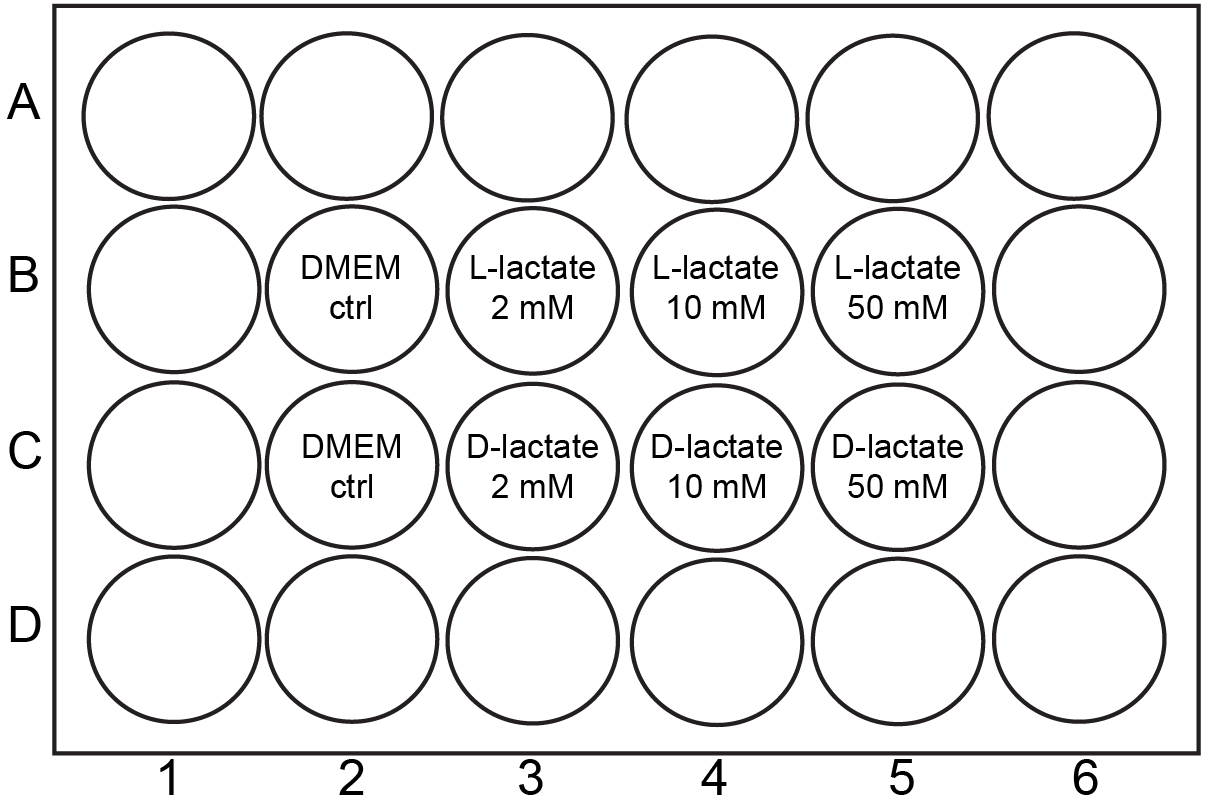
Figure 1. Schematic drawing of experimental setup in the multiwell plate. Initially, bacterial culture is added to all relevant wells and incubated for 3 h under a microscope. After the incubation, lactate solutions are added to wells according to the scheme and the time-lapse started.
- The Axio Observer microscope should be turned on and pre-warmed to 37 °C at least 30 min prior to your experiment. The CO2 should be set to 5% before inserting the multiwell plate to the microscope.
- Preparation during the 3 h incubation
- Perform serial dilutions and plating of the bacterial culture.
Note: Immediately after the 3 h incubation starts, the viable count (108 cfu/ml) of the bacterial culture has to be confirmed by performing serial dilutions and plating. - Dilute the bacterial solution (OD600 = 0.36) by 105 (five 10x dilutions, 50 µl:450 µl) in duplicates and plate 100 µl (10x dilution) of the last dilution to GC plates.
- Incubate the GC plates in a 5% CO2 incubator at 37 °C overnight.
- Count colonies and calculate the viable count (cfu/ml).
- Prepare the microscope.
- In AxioVision software, open multidimensional acquisition from the acquisition on the main tab. In the multidimensional window, click the experiment tab and choose time-lapse and position-list (Figure 2A). Choose a name for your experiment. Settings for experiment should be set to brightfield (BF). The Colibri, which is used for fluorescent microscopy only, should always be turned off and the halogen lamp (HAL), which is used for BF, should be turned off after the experiment.
- In the C tab (channel) choose brightfield in the Dye section and make sure that BF is turned on during acquisition and HAL off after acquisition. Set focus to current (Figure 2B).
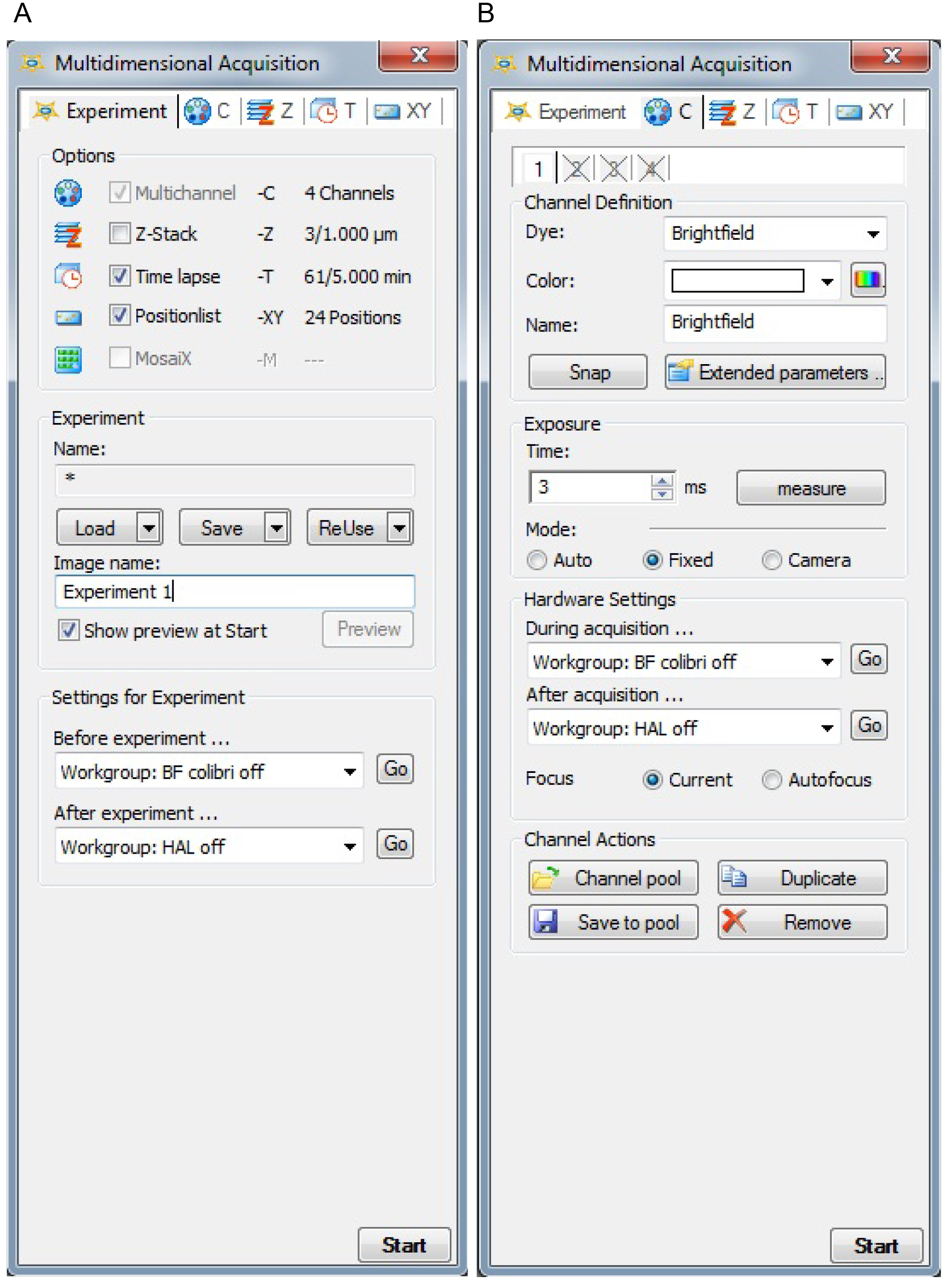
Figure 2. Settings in the AxioVision multidimensional acquisition experiment (A) and channel (B) tabs
- In the T tab (time) choose time-lapse and interval (Figure 3A). To follow the effect on microcolony dispersal, set the multidimensional acquisition to capture image every 5 or 10 min for 5 h total (8 h total incubation).
- In the XY tab, choose both position-list and apply settings before/after timepoint per position (Figure 3B). Click Mark Find and a new window will open. Choose a 24-well plate from the slide/dish collection. Choose the first well containing bacterial solution, well B2 according to the scheme in Figure 1. Click on the live button on the main tab or through acquisition on the main tab. By using bright field illumination and a 40x lens, adjust the focus and set three random positions per well. Place the positions not too close to each other and not too close to the edge of the well, see graphic selected well in Figure 4. Save your positions.
- Start preparing and pre-warming lactate solutions (see Recipes) at different concentrations in DMEM 1 h before addition to microcolonies. The prepared lactate solutions can be kept either in 37 °C incubator or a heating block until addition to microcolonies. For a control, pre-warm DMEM as well.
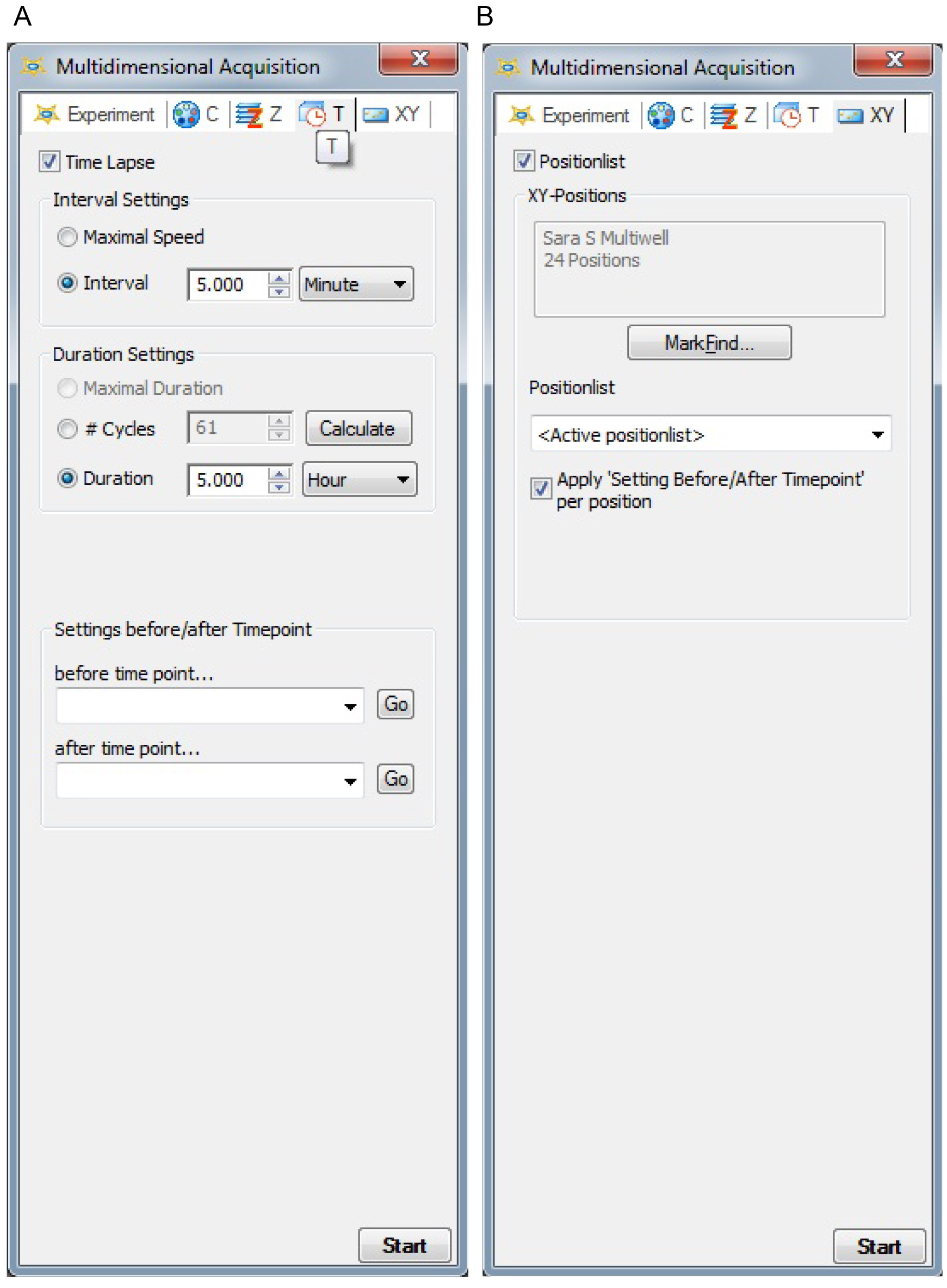
Figure 3. Settings in the AxioVision multidimensional acquisition time (A) and xy (B) tabs
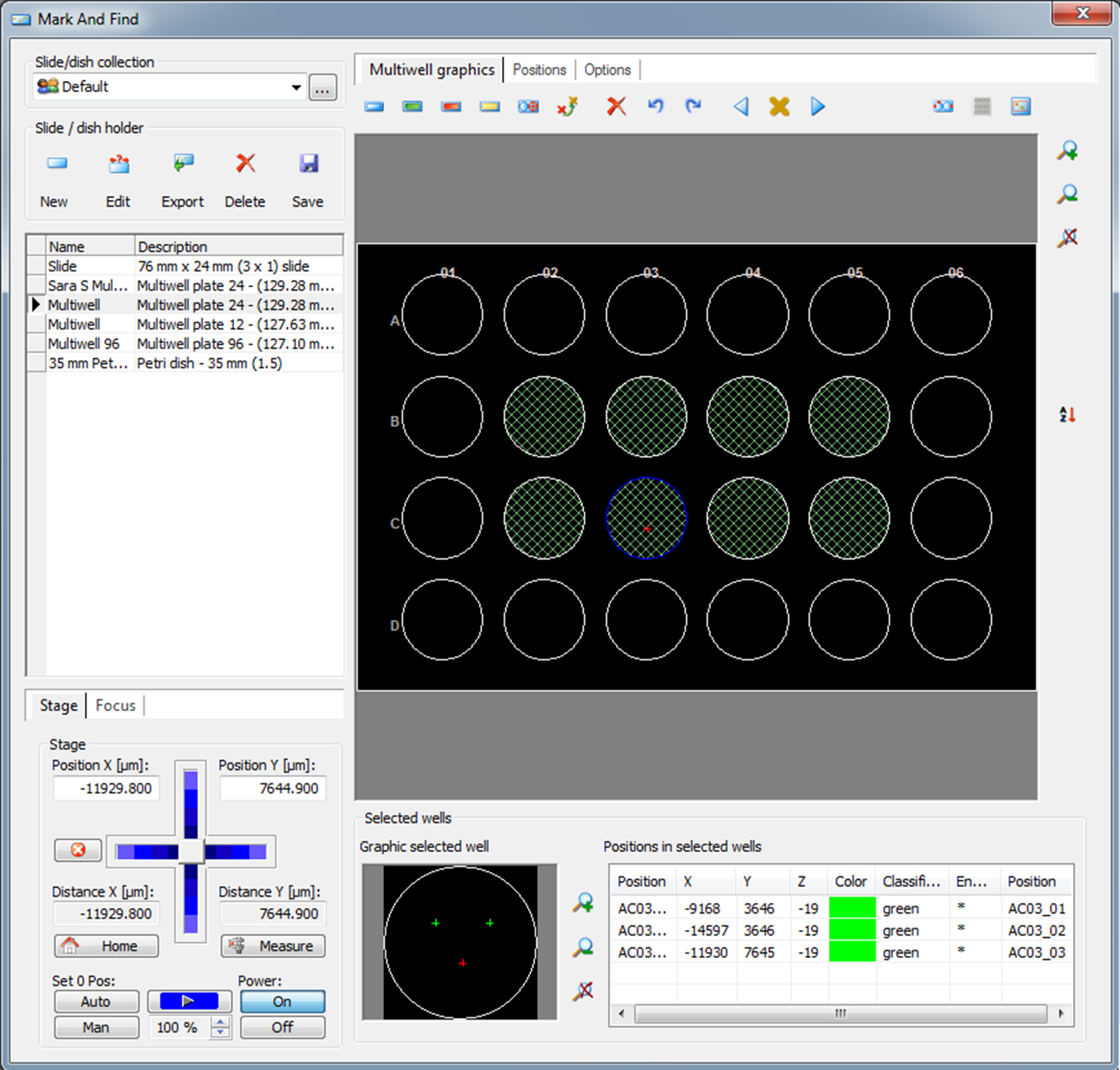
Figure 4. Mark Find window in AxioVision. The image shows how three random positions within a well should be located.
- Perform serial dilutions and plating of the bacterial culture.
- Induction assay: addition of lactate to microcolonies and time-lapse imaging
- After 3 h incubation, open the incubator unit and add 1 ml of each lactate solutions and DMEM control to relevant wells (see scheme in Figure 1). Note that the solutions have to be added quickly but also carefully, so the microcolonies will not be flushed away from the position chosen. Close the incubator unit.
- After all the solutions have been added to relevant wells, quickly examine the positions chosen in the microscope and make sure that the microcolonies chosen have not moved out of position upon addition of lactate solutions. If they do not contain microcolonies, quickly relocate the position. The exposure time needs to be adjusted before starting time lapse. All the adjustments have to be done within 10 min so the time-lapse can be started exactly at 10 min after addition of lactate.
- Start image acquisition, at every 5 or 10 min for 5 h. To avoid differences in timing, capture images in the same order as lactate was added to the wells. Analyze the time-lapse with either the AxioVision software or the Fiji software (see Data analysis).
Data analysis
Acquired images are either analyzed with the software AxioVision or with the license free alternative Fiji (Image J). In Fiji, the files should be opened in XYCZT hyperstack. Evaluate and write down the timing of dispersal phase for every position within the samples, which is the first frame when bacteria start detaching from microcolonies and the last frame when microcolonies are present. If the time-lapse starts exactly 10 min after addition of lactate then the first frame (T1) will be 190 min (3 h and 10 min) and T2 will be 195 min or 200 min depending on if images have been acquired every 5 min or 10 min (Figure 5).
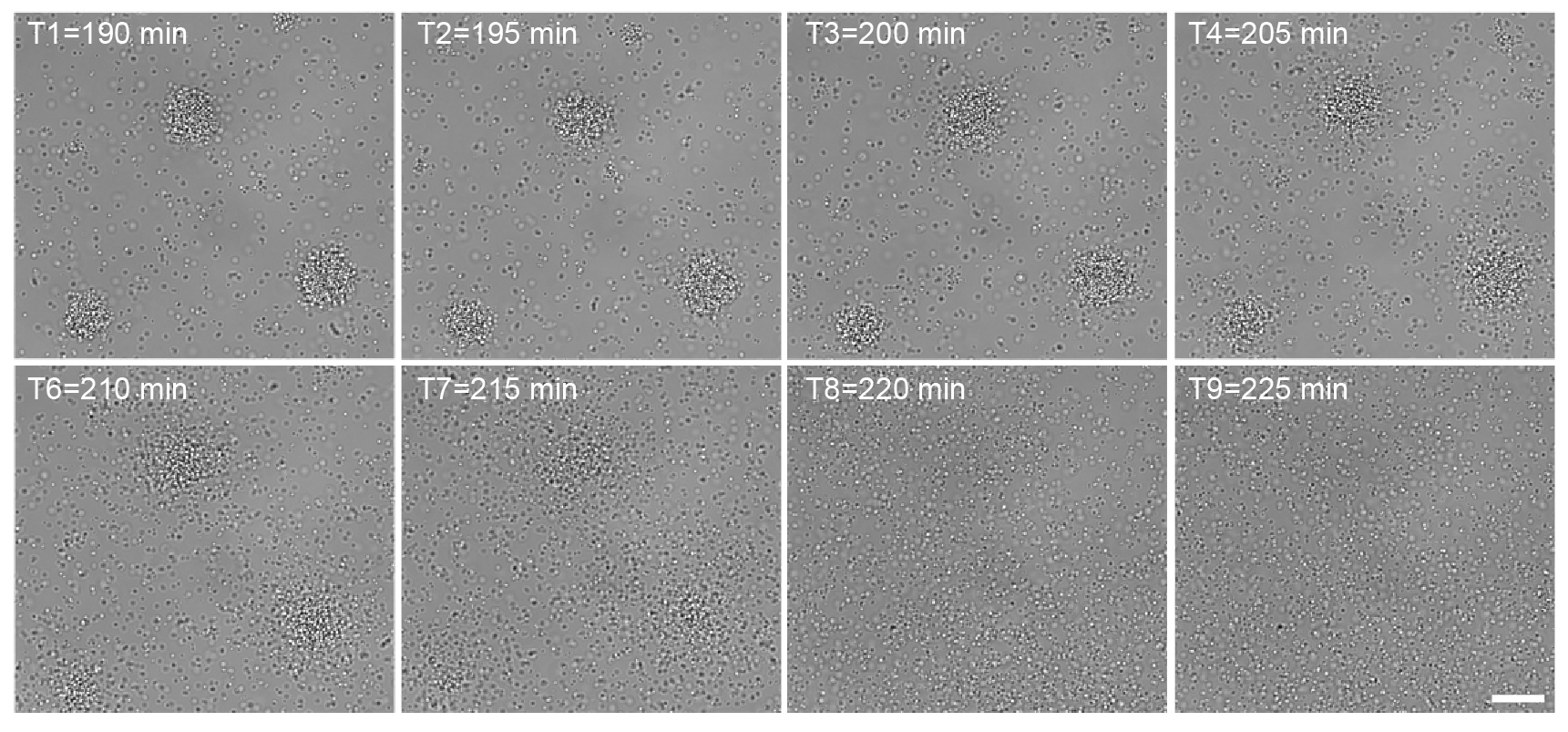
Figure 5. Example of image analysis. In the following time-lapse, 10 mM L-lactate has been added to the well after a 3 h incubation of bacteria. The time-lapse was started 10 min after the addition of lactate (190 min). In frame T3, detachment of bacteria from microcolonies is visible. In frame T8, no aggregates are present anymore. Note that the frames have been cropped in order to create the figure. Scale bar = 20 μm.
- In Excel, make a table with the timing of aggregation phase and dispersal phase for every position within the samples. Aggregation phase is the time from the start of the incubation (0 h) until the last frame before dispersal from microcolonies starts. To get the exact length in time of dispersal phase, subtract the timing when dispersal ends with the timing when dispersal starts. Calculate the average of the three positions. Convert the minutes to hours by dividing the minutes by 60. For length of time of planktonic phase, subtract 8 h (total hours) by length of time in aggregation phase and dispersal phase. Use stacked column chart in Excel to make a graph. According to the example in Figure 5, the dispersal phase occurs between 200 and 220 min, aggregation phase will therefore be from 0-200 min. Planktonic phase will last from 220 min until the end of the timelapse or 480 min (8 h). However, the planktonic phase should be calculated either from the average of the three positions for a single experiment or the average of three independently performed experiments. Data presentation can be seen in Sigurlásdóttir et al. (2017).
- For lactate induced dispersal, three independently performed experiments are sufficient for statistical analysis. However, the number of replicates required to obtain statistically significant data might increase depending on the molecule tested and/or other variabilities in the experimental settings and procedure.
Recipes
- GC agar plates (1 L)
- Add 36 g GC agar medium base to a 1 L bottle
- Add 600 ml of distilled H2O and autoclave at 120 °C for 20 min
- Add up to 1 L of sterile cold distilled H2O
- When the temperature is approximately 60 °C, add 10 ml of Kellogg’s supplement (Recipe below)
- Pour into Petri dishes and allow the plates to cool down and store at 4 °C
- Add 36 g GC agar medium base to a 1 L bottle
- 0.2% Cocarboxylase solution
0.02 g in 100 ml of distilled water
Sterile filter with 0.22 µm filter
Store at 4 °C
- Kellogg’s supplement (200 ml)
- Add 80 g of D-glucose slowly to 100 ml of distilled H2O and allow it to dissolve
- Add 1 g of L-glutamine, 0.1 g of ferric nitrate nonahydrate (FeNO3·9H2O) and 2 ml of 0.2% Cocarboxylase solution (Recipe 2)
- When everything is dissolved, adjust the volume to 200 ml with distilled H2O
- Sterile filter with 0.22 µm filter and store at 4 °C
- Add 80 g of D-glucose slowly to 100 ml of distilled H2O and allow it to dissolve
- 2.5 mM glucose
45.04 g glucose in 100 ml of distilled water
Sterile filter with 0.22 µm filter
Store at 4 °C
- DMEM
For a 500 ml flask of DMEM add the following:
5 ml of GlutaMAX
5 ml of 2.5 mM glucose (Recipe 4)
5 ml of sodium pyruvate
- DMEM/1%FBS
For every 30 ml of DMEM add 0.3 ml of FBS
- Lactate solutions
Notes:- L-lactate and D-lactate solutions are prepared in the same way.
- The lactate solutions are added to the bacterial culture at 1:1 ratio. Therefore the concentration of solutions prepared has to be twice the final concentration.
4 mM (final concentration 2 mM): add 4 µl of 1 M lactate solution to DMEM at a final volume of 1 ml
20 mM (final concentration 10 mM): add 20 µl of 1 M lactate solution to DMEM at a final volume of 1 ml
100 mM (final concentration 50 mM): add 100 µl of 1 M lactate solution to DMEM at a final volume of 1 ml
- L-lactate and D-lactate solutions are prepared in the same way.
Acknowledgments
The work was adapted from a protocol previously published in Sigurlásdóttir et al. (2017). We thank Dr. Sunil D. Saroj for comments on the manuscript. This work was funded by the Swedish Research Council (Dnr 2006-4112, 2012-2415, 2013-2434), The Swedish Cancer Society and Torsten Söderbergs Stiftelse. The authors declare no conflicts of interest.
References
- Engman, J., Negrea, A., Sigurlásdóttir, S., Georg, M., Eriksson, J., Eriksson, O. S., Kuwae, A., Sjolinder, H. and Jonsson, A. B. (2016). Neisseria meningitidis polynucleotide phosphorylase affects aggregation, adhesion, and virulence. Infect Immun 84(5): 1501-1513.
- Eriksson, J., Eriksson, O. S. and Jonsson, A. B. (2012). Loss of meningococcal PilU delays microcolony formation and attenuates virulence in vivo. Infect Immun 80(7): 2538-2547.
- Pujol, C., Eugene, E., de Saint Martin, L. and Nassif, X. (1997). Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect Immun 65(11): 4836-4842.
- Pujol, C., Eugene, E., Marceau, M. and Nassif, X. (1999). The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc Natl Acad Sci U S A 96(7): 4017-4022.
- Rahman, M., Kallstrom, H., Normark, S. and Jonsson, A. B. (1997). PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol Microbiol 25(1): 11-25.
- Sigurlásdóttir, S., Engman, J., Eriksson, O. S., Saroj, S. D., Zguna, N., Lloris-Garcera, P., Ilag, L. L. and Jonsson, A. B. (2017). Host cell-derived lactate functions as an effector molecule in Neisseria meningitidis microcolony dispersal. PLoS Pathog 13(4): e1006251.
- Stephens, D. S. (2009). Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 27 Suppl 2: B71-77.
- Trivedi, K., Tang, C. M. and Exley, R. M. (2011). Mechanisms of meningococcal colonisation. Trends Microbiol 19(9): 456-463.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sigurlásdóttir, S., Eriksson, O. S., Eriksson, J. and Jonsson, A. (2018). Live-cell Imaging of Neisseria meningitidis Microcolony Dispersal Induced by Lactate or Other Molecules. Bio-protocol 8(2): e2695. DOI: 10.21769/BioProtoc.2695.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial cell biology > Cell imaging
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









