- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visible Immunoprecipitation (VIP) Assay: a Simple and Versatile Method for Visual Detection of Protein-protein Interactions
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2687 Views: 13609
Reviewed by: Alessandro DidonnaLai-Yee WongMirko Messa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 287 Views

Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 443 Views
Abstract
The visible immunoprecipitation (VIP) assay is a convenient alternative to conventional co-immunoprecipitation (Katoh et al., 2015). By processing lysates from cells co-expressing GFP-fusion and RFP-fusion proteins for immunoprecipitation with GST-tagged anti-GFP Nanobody and glutathione-Sepharose beads, protein-protein interactions can be visualized by directly observing the beads bearing immunoprecipitates under a fluorescence microscope. This assay can examine a large number of protein combinations at one time, without requiring time-consuming procedures, including SDS-PAGE and immunoblotting. Furthermore, the VIP assay can examine complicated one-to-many and many-to-many protein interactions. Another important point of the VIP assay is the use of nanobodies for immunoprecipitation. A Nanobody is a single-domain antibody derived from Camelidae (camels and relatives). Because of its small size, high-affinity, high-specificity, and stability, anti-GFP Nanobody expressed in E. coli can be purified on a large scale, and used virtually inexhaustibly for immunoprecipitation experiments. Here we describe protocols for preparation of GST-tagged anti-GFP Nanobody and the VIP assay.
Keywords: Visible immunoprecipitation (VIP)Background
Almost all proteins in cells function by interacting with other proteins. Revealing the protein-protein interaction network is the key to understand the functions of the proteins. Various methods such as yeast two-hybrid system, GST pull-down, and co-immunoprecipitation have been developed to analyze protein-protein interactions. Recently, we have developed a new method for protein-protein interaction analysis called visible immunoprecipitation (VIP) assay (Katoh et al., 2015). The most important advantage of VIP assay is that it is handy and convenient. This assay can examine a large number of protein combinations in a short time, without requiring time-consuming procedures, including SDS-PAGE and immunoblotting. Furthermore, the VIP assay can determine interactions between more than two proteins at a time. This powerful tool can be used to reveal the intricate architectures of multi-protein complexes, which cannot be determined by conventional protein-protein interaction assays. By taking advantage of the VIP assay, we have elucidated architectures of multi-subunit complexes, the BBSome (composed of 8 subunits) (Katoh et al., 2015), IFT-B (16 subunits) (Katoh et al., 2016), and IFT-A (6 subunits) (Hirano et al., 2017), all of which are involved in protein trafficking within the cilia.
Materials and Reagents
- 10 ml test tubes (ASIA KIZAI, catalog numbers: 2221C010B-10 )
- 50 ml tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339652 )
- 15 ml tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339650 )
- 6-well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 140675 )
- 1.5 ml tubes (BIO-BIK, catalog number: RC-0150 )
- 0.2 ml 8-Tube Strips (Greiner Bio One International, catalog numbers: 673210 and 373270 )
- 96-well plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 167008 )
- 0.22 µm PVDF filter (Millex-GV 0.22 µm PVDF 33 mm Gamma Sterilized) (Merck, catalog number: SLGV033RS )
- pGEX6P1-GFP-Nanobody (Addgene, catalog number: 61838 )
- Escherichia coli BL21-CodonPlus(DE3)-RIPL strain (Agilent Technologies, catalog number: 230280 )
- HEK293T cells (e.g., ATCC, catalog number: CRL-3216 )
- Fluorescent protein expression vectors (e.g., mEGFP-C1/N1 [Addgene, catalog numbers: 54759 and 54767 ], pmCherry-C1/N1 [Takara Bio, Clontech, catalog numbers: 632524 and 632523 ], or pTagRFP-C/N [Evrogen, catalog numbers: FP141 and FP142 ])
- Luria-Bertani (LB) medium (NACALAI TESQUE, catalog number: 20068-75 )
- LB agar plates (NACALAI TESQUE, catalog number: 20069-65 )
- Ampicillin (NACALAI TESQUE, catalog number: 02739-32 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (NACALAI TESQUE, catalog number: 19742-94 )
- Polyethylene Glycol Mono-p-isooctylphenyl Ether (Triton X-100) (NACALAI TESQUE, catalog number: 12967-45 )
- Glutathione-Sepharose 4B (GE Healthcare, catalog number: 17075601 )
- Phosphate buffered saline without Ca2+ and Mg2+ [PBS(-)] (NACALAI TESQUE, catalog number: 1148215 )
- CBB Stain One Super (NACALAI TESQUE, catalog number: 11642-31 )
- Dulbecco’s modified Eagle medium (DMEM), high glucose (NACALAI TESQUE, catalog number: 08458-16 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- Opti-MEM (Thermo Fisher Scientific, catalog number: 31985070 )
- Dithiothreitol (DTT) (NACALAI TESQUE, catalog number: 14112-52 )
- Polyethyleneimine (PEI) Max (Mw 40,000) (Polysciences, catalog number: 24765-2 )
- HEPES (NACALAI TESQUE, catalog number: 17546-05 )
- Sodium chloride (NaCl) (NACALAI TESQUE, catalog number: 31320-05 )
- Glycerol (NACALAI TESQUE, catalog number: 17018-25 )
- Protease Inhibitor Cocktail for general use (100x) (NACALAI TESQUE, catalog number: 04080-11 )
Note: This protease inhibitor cocktail contains 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), aprotinin, E-64, leupeptin hemisulfate monohydrate, and disodium dihydrogen ethylenediaminetetraacetate dihydrate (EDTA). - Bovine serum albumin (BSA) (NACALAI TESQUE, catalog number: 08587-84 )
- Protease Inhibitor Cocktail for use with mammalian cell and tissue extracts (100x) (NACALAI TESQUE, catalog number: 25955-11 )
Note: This protease inhibitor cocktail contains 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), aprotinin, E-64,leupeptin hemisulfate monohydrate, bestatin, and pepstatin A. - Binding buffer (see Recipes)
- Washing buffer (see Recipes)
- 2 mg/ml PEI Max reagent (see Recipes)
- HEPES, NaCl, Triton X-100, and glycerol (HNTG) buffer (see Recipes)
Equipment
- Multichannel pipette (20-200 µl) (Greiner Bio One International, catalog number: 89008200 )
- Aspirator
- Large centrifuge (Hitachi Koki, model: CR22G )
- Cooling centrifuge that can be set at 4 °C (TOMY SEIKO, model: EX-136 )
- Desktop centrifuge (Eppendorf, model: 5415 R )
- Centrifuge for PCR tube (BM Equipment, model: ForceMini )
- Shaking incubator (TAITEC, model: BR-23FP )
- Rotator (TAITEC, model: RT-50 )
- Sonicator (Misonix, model: S-4000 )
- Fluorescence microscope (Keyence, model: BZ-8000 )
- Objective lens (Nikon, PlanApo 20x NA0.75 WD1.00)
- 500 ml flask (AGC Techno Glass, catalog number: 4551FK500R )
- Spectrophotometer (GE healthcare, model: UltrospecTM 2100 pro )
- Cuvette (GE Healthcare, catalog number: 80-2076-38 )
Procedure
- Preparation of GST-tagged anti-GFP Nanobody
Here we introduce the method for preparing GST-tagged anti-GFP Nanobody. Under our conditions, we usually purify approximately 1 mg of the protein from 200 ml of E. coli culture.- Pick up a single colony of E. coli carrying pGEX6P1-GFP-Nanobody, and inoculate a starter culture of 3 ml of LB medium plus 50 µg/ml ampicillin in a test tube with the colony, and incubate it overnight at 37 °C with vigorous shaking at 150-200 rpm.
- Inoculate a flask containing 200 ml of LB medium plus 50 µg/ml ampicillin with 2 ml of the overnight culture, and incubate it at 37 °C with shaking at 130 rpm until the culture reaches OD600 = 0.5. (It takes 2-4 h.)
- To induce expression of GST-tagged anti-GFP Nanobody, add 200 µl of 100 mM IPTG (to a final concentration 0.1 mM) to the flask, and incubate the flask for 4 h at 30 °C with shaking at 130 rpm.
- Transfer the culture to an appropriate centrifuge tube, and harvest the cells by centrifugation at 5,550 x g (e.g., 6,000 rpm in a Himac CR22G centrifuge) for 15 min at 4 °C.
- Pour away the supernatant and suspend the cells in 10 ml of binding buffer (see Recipes) by vortexing.
- After transferring the cell suspension to a 50 ml tube, disrupt the cells by sonication. Sonication is performed on ice for 5 rounds (15 sec ON and 15 sec OFF) at 30% amplitude using Astrason S4000.
- Add 0.5 ml of 20% Triton X-100 diluted in H2O to the tube to a final concentration of 1%, and incubate it for 20 min at 4 °C (or on ice) to help cell lysis.
- Centrifuge the tube at 20,700 x g (e.g., 12,000 rpm in a Himac CR22G centrifuge) for 20 min at 4 °C, and transfer the supernatant to a 15 ml tube.
Note: We usually divide the supernatant into 2 x 5 ml. Hereafter we use 2 tubes. - Add 2.5 ml (bed volume) of glutathione-Sepharose 4B beads equilibrated in PBS(-) to 5 ml of the supernatant, and allow binding of the Nanobody to the beads by rotating the tube overnight at 4 °C (12-24 h).
- Centrifuge the tube at 780 x g (e.g., 2,000 rpm in an EX-136 centrifuge) for 2 min at 4 °C, and remove the supernatant by aspiration.
- Add 10 ml of ice-cold washing buffer (see Recipes) to the precipitate.
- Repeat Steps A10 and A11 eight times.
- Keep the beads as a 1:1 slurry in the washing buffer (namely, 2.5 ml of the beads and 2.5 ml of the washing buffer). The beads can be stored for several months at 4 °C.
Note: We usually divide the beads slurry into 1 ml (bed volume 500 µl) aliquots. - Check the quality and estimate the quantity of purified GST-tagged anti-GFP Nanobody by SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining. Under our preparation conditions, approximately 1 µg/5 µl (bed volume of beads) of the Nanobody protein is purified. A typical result is shown in Figure 1.
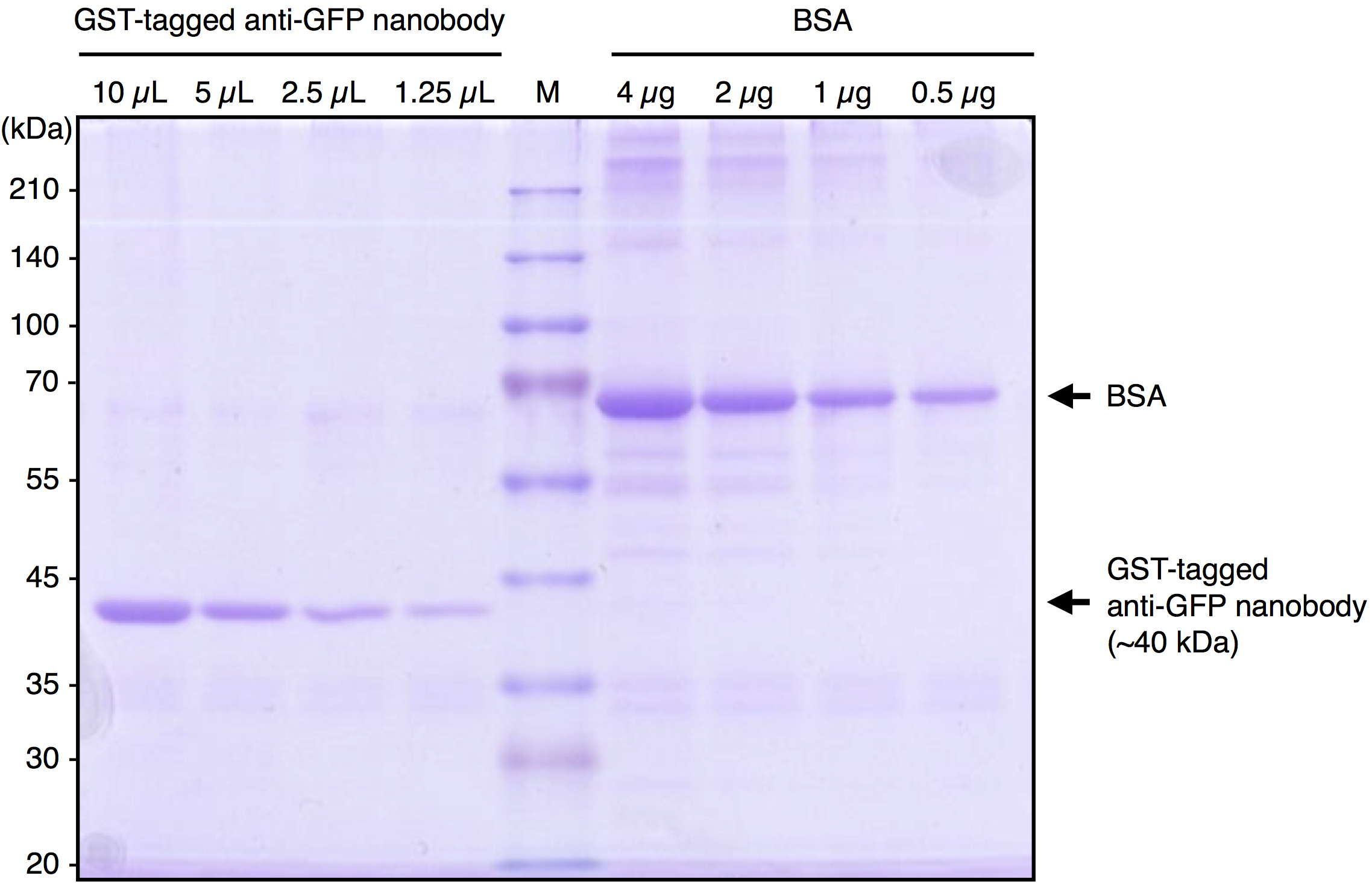
Figure 1. A typical result of purified GST-tagged anti-GFP Nanobody. Purified GST-tagged anti-GFP Nanobody and BSA were subjected to SDS-PAGE followed by CBB staining. By comparing the band intensity of the purified protein with that of BSA, the concentration of purified GST-tagged anti-GFP Nanobody was estimated to be approximately 1 μg/5 μl (bed vol.).
- Pick up a single colony of E. coli carrying pGEX6P1-GFP-Nanobody, and inoculate a starter culture of 3 ml of LB medium plus 50 µg/ml ampicillin in a test tube with the colony, and incubate it overnight at 37 °C with vigorous shaking at 150-200 rpm.
- VIP assay
Here we introduce how to perform the VIP assay (Figure 2 and Video 1).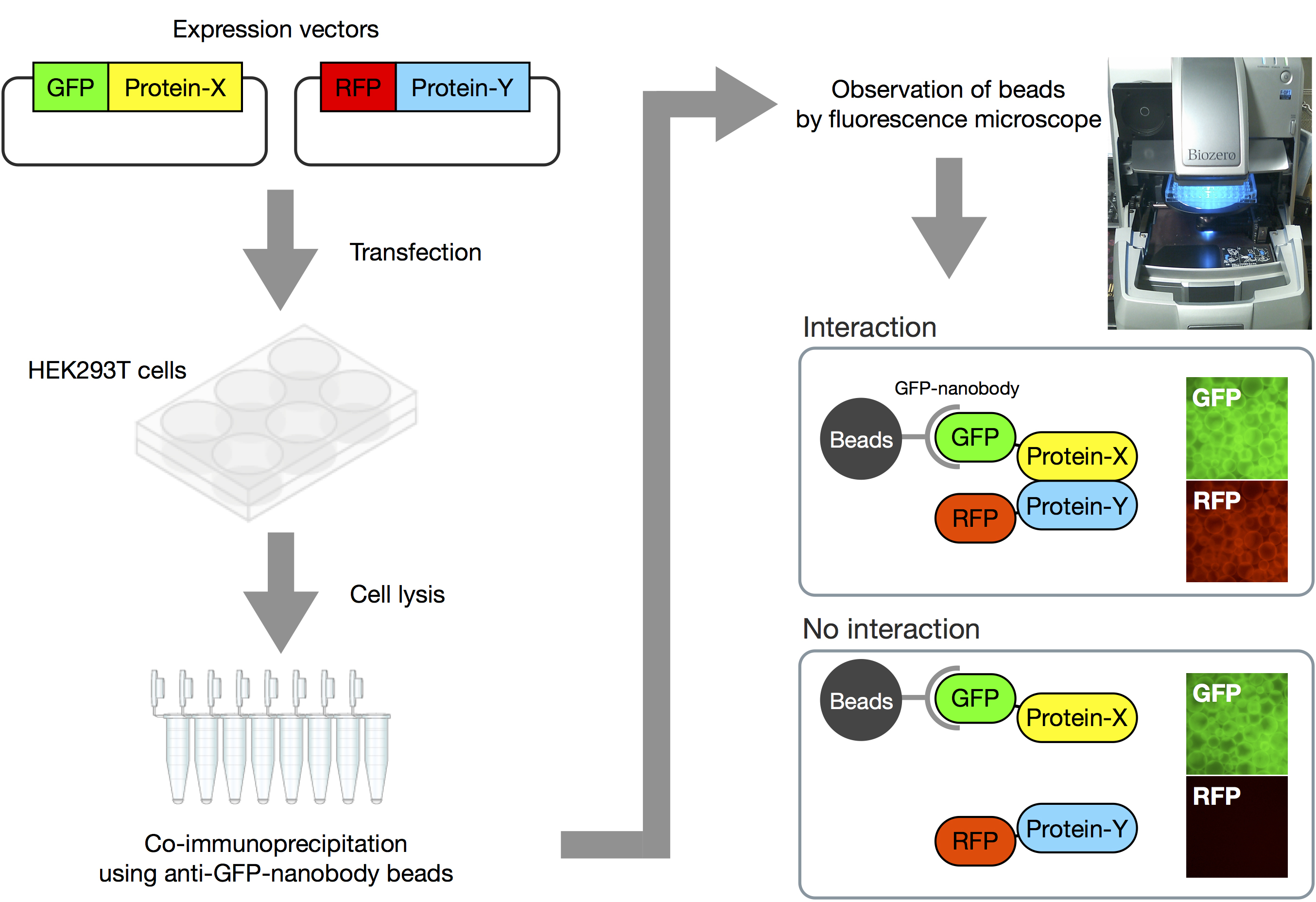
Figure 2. Overview of VIP assay. Expression vectors for GFP-fusion and RFP-fusion proteins are co-transfected into HEK293T cells. After 24 h, expression of fluorescent proteins is confirmed by fluorescence microscopy. Next, lysates of the transfected cells are prepared, and processed for immunoprecipitation using GST-tagged anti-GFP Nanobody beads. Finally, the beads bearing immunoprecipitates are observed under a fluorescence microscope. When two proteins interact with each other, both GFP and RFP signals are detectable on the beads. On the other hand, when the two proteins do not interact, only GFP signals are observed.Video 1. Demonstration of VIP assay. The video shows how to perform the VIP assay.- Seed HEK293T cells (approximately 8 x 105 cells) into a 6-well plate and incubate them for 24 h in 2 ml of DMEM high glucose supplemented with 5% FBS.
- Co-transfect the cells with expression vectors for GFP-fusion and RFP-fusion proteins using a PEI Max reagent (see Recipes).
- Mix 250 µl of Opti-MEM with 4 µg of DNA in a 1.5 ml tube (e.g., 2 μg of a GFP expression vector and 2 μg of an RFP expression vector).
- Mix 250 µl of Opti-MEM with 10 µl of 2 mg/ml PEI Max reagent in another 1.5 ml tube, and incubate for 5 min at room temperature.
- Mix the DNA solution with the PEI Max solution by vortexing, and incubate it for 20 min at room temperature.
- Add 500 µl of the DNA/PEI mixture to each well of the 6-well plate.
- Mix 250 µl of Opti-MEM with 4 µg of DNA in a 1.5 ml tube (e.g., 2 μg of a GFP expression vector and 2 μg of an RFP expression vector).
- After 24 h, check the expression of GFP-fusion and RFP-fusion proteins by observing the cells under a fluorescence microscope.
- Remove completely the medium from the plate by aspiration, add 250 µl of ice-cold HNTG buffer (see Recipes) to each well, and incubate the plate for 5-10 min on ice to detach adherent cells.
- Suspend the cells by pipetting, transfer the cell suspension to a 1.5 ml tube, and incubate the tube for 15 min on ice to lyse the cells.
- Centrifuge the tube at 16,100 x g (e.g., 13,200 rpm in a 5415 R centrifuge) at 4 °C for 15 min.
- Transfer 200 µl of the supernatant to 0.2 ml 8-Tube Strips containing GST-tagged anti-GFP Nanobody prebound to glutathione-Sepharose beads (approximately 5 µl (bed volume) of the beads), and incubate the 8-Tube Strips for 1 h at 4 °C with constant rotation using a tube rotator.
- Centrifuge the 8-Tube Strips for 30 sec, and remove the supernatant by aspiration.
- Add 180 µl of ice-cold HNTG buffer, and mix the content by tapping the 8-Tube Strips.
- Repeat Steps B8 and B9 three times.
- Finally, suspend the beads in 180 µl of ice-cold PBS(-), and transfer the beads suspension to a 96-well plate.
- Observe green and red fluorescence on the beads under a fluorescence microscope, and acquire images. To compare the fluorescence intensity, images are acquired under fixed conditions (exposure time and ISO sensitivity of the camera).
- Collect the beads if immunoblotting is required.
- Seed HEK293T cells (approximately 8 x 105 cells) into a 6-well plate and incubate them for 24 h in 2 ml of DMEM high glucose supplemented with 5% FBS.
Data analysis
- Examples of VIP assay
The intraflagellar transport (IFT)-B complex, which is responsible for ciliary protein trafficking is a huge protein complex consisting of 16 subunits. We have delineated the architecture of the IFT-B complex by determining not only binary interactions but also one-to-many and many-to-many protein interactions of IFT-B subunits by the VIP assay (Katoh et al., 2016). Here we show examples of interaction analyses of some IFT-B subunits.- Experimental example 1: binary protein interaction
The VIP assay was performed on GFP-tagged and mCherry(mChe)-tagged IFT74 and IFT81 (Figure 3A). The red signals in the fourth and fifth panels from the left indicate that IFT74 and IFT81 interact with each other. - Experimental example 2: one-to-two protein interaction
The VIP assay was performed on GFP-tagged IFT22 and mChe-tagged IFT74, IFT81, or IFT74 + IFT81 (Figure 3B). GFP-IFT22 did not interact with either mChe-IFT74 or mChe-IFT81 alone. However, GFP-IFT22 interacted with mChe-tagged IFT74 + IFT81 heterodimer (rightmost panel in Figure 3B). To confirm that both IFT74 and IFT81 are bound to IFT22, immunoblotting was performed after the VIP assay (Figure 3C). The results of the VIP assay and immunoblotting show that IFT22 interact with only the IFT74-IFT81 heterodimer (Figure 3D).
As described above, the VIP assay is a versatile and flexible method that can comprehensively examine binary protein interactions and examine more complicated one-to-many and many-to-many protein interactions.
Finally, we describe notes about the VIP assay. In this method, it is very important to use appropriate positive and negative controls. As a positive control, proteins already known to interact should be used. As a negative control, only fluorescent proteins are used. Using appropriate experimental controls facilitates determining whether protein-protein interactions are positive or negative.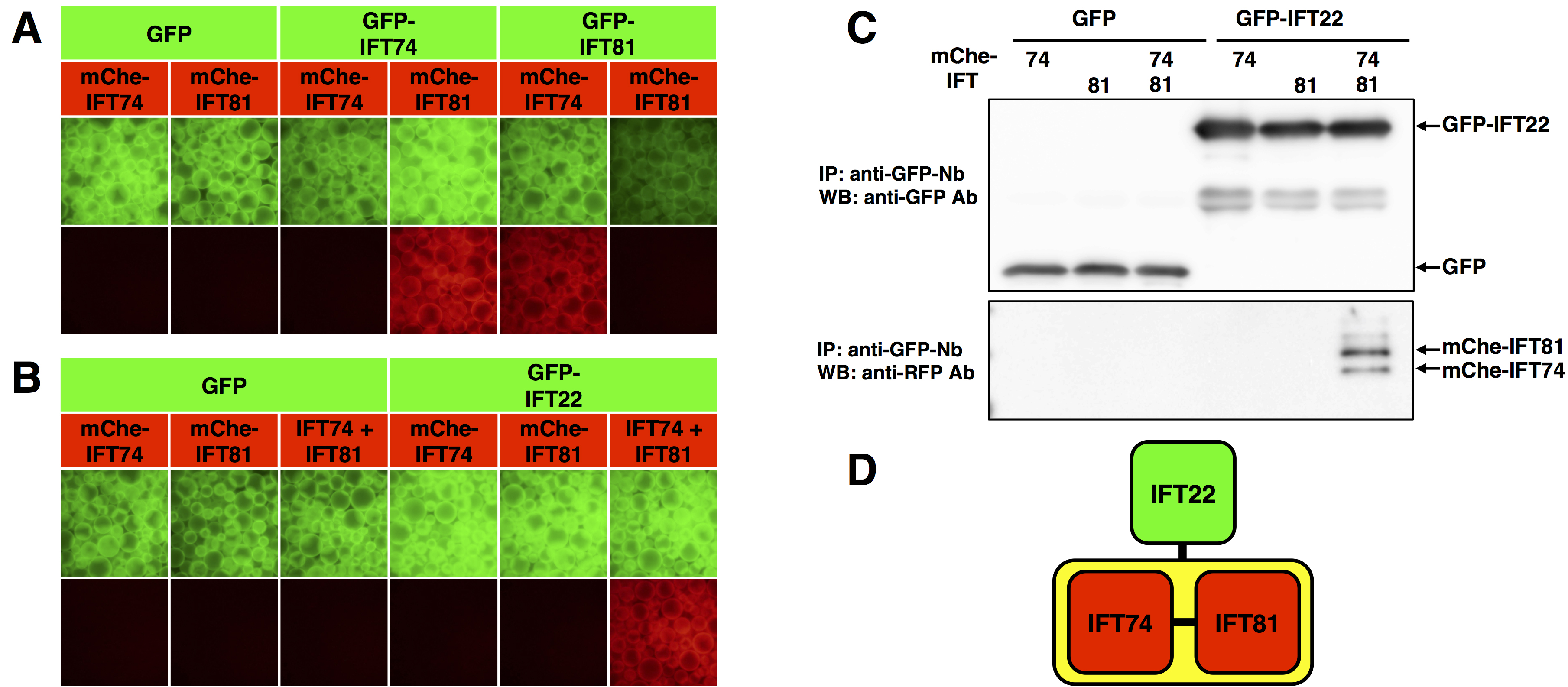
Figure 3. Interactions among IFT subunits revealed by VIP assay. A. Binary protein interaction. The VIP assay was performed on GFP- or mChe-tagged IFT74 and IFT81. B. One-to-two protein interaction. The VIP assay was performed on GFP-tagged IFT22 and mChe-tagged IFT74, IFT81, or IFT74 + IFT81. C. Immunoblotting was performed after the VIP assay shown in B. D. The interaction mode of IFT22, IFT74, and IFT88. (From Katoh and Nakayama, 2017; adapted with permission from YODOSHA Co., Ltd.)
- Experimental example 1: binary protein interaction
Recipes
- Binding buffer
PBS(-)
5 mM DTT
Protease Inhibitor Cocktail for general use (0.5 mM AEBSF, 0.15 µM aprotinin, 1 µM E-64, 1 µM leupeptin hemisulfate monohydrate, and 0.5 mM EDTA) - Washing buffer
PBS(-)
5 mM DTT
0.1% Triton X-100 - 2 mg/ml PEI Max reagent
20 mg of PEI Max is dissolved in 10 ml of Milli-Q grade water sterilized by filtration
The solution is divided into 1 ml aliquots, and can be stored at 4 °C for several months
For more long-term, it can be stored at -20 °C - HEPES, NaCl, Triton X-100, and glycerol (HNTG) buffer
Note: This is an example of lysis buffer. Other lysis buffers can also be used for VIP assay.
20 mM HEPES (pH7.4)
150 mM NaCl
0.1% (w/v) Triton X-100
10% (w/v) glycerol
Protease Inhibitor Cocktail for use with mammalian cell and tissue extracts (1 mM AEBSF, 0.8 µM aprotinin, 15 µM E-64, 20 µM leupeptin hemisulfate monohydrate, 50 µM bestatin, and 10 µM pepstatin A)
Acknowledgments
This protocol was adapted from and used in Katoh et al. (2015), Katoh et al. (2016), and Katoh and Nakayama (2017). This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas ‘Cilia and Centrosome’ from the Ministry of Education, Culture, Sports, Science and Technology, Japan (grant number 15H01211 to Ka.N.); grants from the Japan Society for the Promotion of Science (grant numbers 15H04370 to Ka.N., and 15K07929 to Y.K.); and from the Takeda Science Foundation and the Uehara Memorial Foundation to Y.K. The authors declare no conflicts of interest.
References
- Hirano, T., Katoh, Y. and Nakayama, K. (2017). Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell 28(3): 429-439.
- Katoh, Y. and Nakayama K. (2017). Visible immunoprecipitation assay: a convenient and versatile method for studying protein‒protein interactions using fluorescent fusion proteins. Experimental Medicine 35: 85-95 (written in Japanese).
- Katoh, Y., Nozaki, S., Hartanto, D., Miyano, R. and Nakayama, K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci 128(12): 2351-2362.
- Katoh, Y., Terada, M., Nishijima, Y., Takei, R., Nozaki, S., Hamada, H. and Nakayama, K. (2016). Overall architecture of the Intraflagellar Transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem 291: 10962-10975.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Katoh, Y., Nakamura, K. and Nakayama, K. (2018). Visible Immunoprecipitation (VIP) Assay: a Simple and Versatile Method for Visual Detection of Protein-protein Interactions. Bio-protocol 8(1): e2687. DOI: 10.21769/BioProtoc.2687.
Category
Neuroscience > Cellular mechanisms > Receptor-ligand binding
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












