- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Bacterial Aggregation Assay in the Presence of Cyclic Lipopeptides
(*contributed equally to this work) Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2686 Views: 9614
Reviewed by: Valentine V TrotterJose Antonio Reyes-DariasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Functional Assay for Measuring Bacterial Degradation of Gemcitabine Chemotherapy
Serkan Sayin and Amir Mitchell
Sep 5, 2023 1741 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3103 Views

Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
Chen Tang [...] Guangxin Xiang
Feb 5, 2025 1985 Views
Abstract
Lipopeptides is an important class of biosurfactants having antimicrobial and anti-adhesive activity against pathogenic bacteria. These include surfactin, fengycin, iturin, bacillomycin, mycosubtilin, lichenysin, and pumilacidin (Arima et al., 1968; Naruse et al., 1990; Yakimov et al., 1995; Steller and Vater, 2000; Roongsawang et al., 2002; Vater et al., 2002). To date, none of these lipopeptides have been reported to possess any anti-motility activity. We isolated, purified and characterized two novel cyclic lipopeptides (CLPs) from Bacillus sp. 176 using high performance liquid chromatography, mass spectrometry and nuclear magnetic resonance spectroscopy. CLPs dramatically suppress the motility of pathogenic bacterium Vibrio alginolyticus 178, and promote cellular aggregation without inducing cell death. Cell aggregation assay was performed with the modification according to methods described by Dalili for anti-biofilm assay (Dalili et al., 2015). In future, this assay can be adapted to test both the cell aggregation and anti-biofilm activity of lipopeptide-like active substances derived from bacteria.
Keywords: Cyclic lipopeptidesBackground
Overuse of broad-spectrum antibiotics and the accompanying proliferation of drug-resistant bacteria have stimulated efforts to develop environment-friendly biocontrol measures to reduce health hazards and environmental pollution (Nam et al., 2016; Sajitha et al., 2016). In recent years, the anti-microbial properties of biological surfactants have been increasingly recognized and harnessed for antibacterial, antifungal, and antiviral applications (Cameotra and Makkar, 2004; Singh and Cameotra, 2004; Rodrigues et al., 2006). Lipopeptides are the most widely reported class of biosurfactants having antimicrobial and 100 anti-adhesive activity against pathogenic bacteria, due to the amphipathic nature of their peptide and fatty acid components (Das et al., 2008; Dalili et al., 2015). In this study, two cyclic lipopeptides (CLPs) derived from a competing bacterium (Bacillus sp. 176) are found to inhibit the motility and promote the aggregation of V. alginolyticus 178. We purified and characterized the active anti-motility compounds and determined their structural and functional properties. In order to explore the mechanism of action of the CLPs, their impact on cell aggregation, adherence, and the expression of flagellar assembly components in V. alginolyticus were also investigated.
Materials and Reagents
- Pipette tips (Corning, Axygen®, catalog numbers: T-200-Y-STK , T-300-L-R , T-1000-B )
- 15 ml culture tube (Bomei, catalog number: SGJS15ML )
- Flat bottom 96-well microtiter plate (Corning, catalog number: 3628 )
- Coverslips (CITOTEST LABWARE MANUFACTURING, catalog number: 80340-1130 )
- Bacterial strains (Identified and stored in our lab)
- V. alginolyticus 178
- V. anguillarum
- V. splendidus
- V. vulnificus
- Pseudomonas aeruginosa
- P. stutzeri
- Staphylococcus aureus
- Bacillus sp. 176
- B. subtilis
- V. alginolyticus 178
- Methanol (Sinopharm Chemical Reagent, catalog number: 10014108 )
- Ethanol (Sinopharm Chemical Reagent, catalog number: 10009259 )
- Peptone (Solarbio, catalog number: P8450 )
- Tryptone (OXOID, catalog number: LP0042 )
- Yeast extract (OXOID, catalog number: LP0021 )
- Agar powder (Solarbio, catalog number: A8190 )
- Sodium chloride (NaCl) (Sinopharm Chemical Reagent, catalog number: 10019318 )
- Crystal violet (Sinopharm Chemical Reagent, catalog number: 71012314 )
- Glacial acetic acid (Sinopharm Chemical Reagent, catalog number: 10000218 )
- Gelatin (Solarbio, catalog number: G8060 )
- 25% glutaraldehyde solution (Sinopharm Chemical Reagent, catalog number: 30092436 )
- Dimethyl sulfoxide (DMSO) (MP Biomedicals, catalog number: 02196055 )
- Saline LB broth (see Recipes)
- LB broth (see Recipes)
- 1% (w/v) solution of crystal violet (see Recipes)
- 30% (v/v) acetic acid (see Recipes)
- Modified 2216E broth (see Recipes)
- 1% (w/v) gelatin solution (see Recipes)
- 5% glutaraldehyde solution (see Recipes)
- Sterile saline solution (see Recipes)
- 10 mg/ml CLPs (see Recipes)
Equipment
- 1-10 μl pipettor (Gilson, model: P10N )
- 20-200 μl pipettor (Gilson, model: P200N )
- 100-1,000 μl pipettor (Gilson, model: P1000N )
- Autoclave sterilizer (Zealway Instrument, model: GI80TW )
- Constant temperature shaker (CRYSTAL, model: IS-RDS3 )
- Centrifuge (Eppendorf, model: 5418 R )
- SYNERGY-H1 microplate reader (BioTek Instruments, model: Synergy H1 )
- Scanning electron microscope (SEM) (Hitachi, model: S-3400N )
- Transmission electron microscope (TEM) (Hitachi, model: H-7650 )
- Biological safety cabinet (Heal Force, model: HFsafe 900LC )
Procedure
- Bacterial strains culture
- Single colonies of bacterial strains used in this protocol, including V. alginolyticus 178, V. anguillarum, V. splendidus, V. vulnificus, Pseudomonas aeruginosa, P. stutzeri, Staphylococcus aureus, Bacillus sp. 176 and B. subtilis, were selected from their pure culture plates.
- V. alginolyticus 178, V. anguillarum, V. splendidus, V. vulnificus, Pseudomonas aeruginosa, P. stutzeri and Bacillus sp. 176 were cultured in 5 ml saline Luria-Bertani (LB) broth (see Recipes) in tubes at 28 °C overnight with shaking at 170 rpm.
- Staphylococcus aureus and B. subtilis were cultured in 5 ml LB broth in tubes at 37 °C overnight with shaking at 170 rpm.
- Single colonies of bacterial strains used in this protocol, including V. alginolyticus 178, V. anguillarum, V. splendidus, V. vulnificus, Pseudomonas aeruginosa, P. stutzeri, Staphylococcus aureus, Bacillus sp. 176 and B. subtilis, were selected from their pure culture plates.
- Aggregation assay of V. alginolyticus 178 in culture tubes
- After overnight culture, dilute the cell suspension of V. alginolyticus 178 at 1:100 with saline LB broth.
- Prepare six sterilized 15 ml culture tubes.
- Add 3 ml bacterial suspension into each tube.
- Treat the cells with 90 μl (10 mg/ml) CLPs with a final concentration of 300 μg/ml or same volume DMSO, respectively, and incubate statically at 28 °C for 24 h after homogenization (Figure 1A).
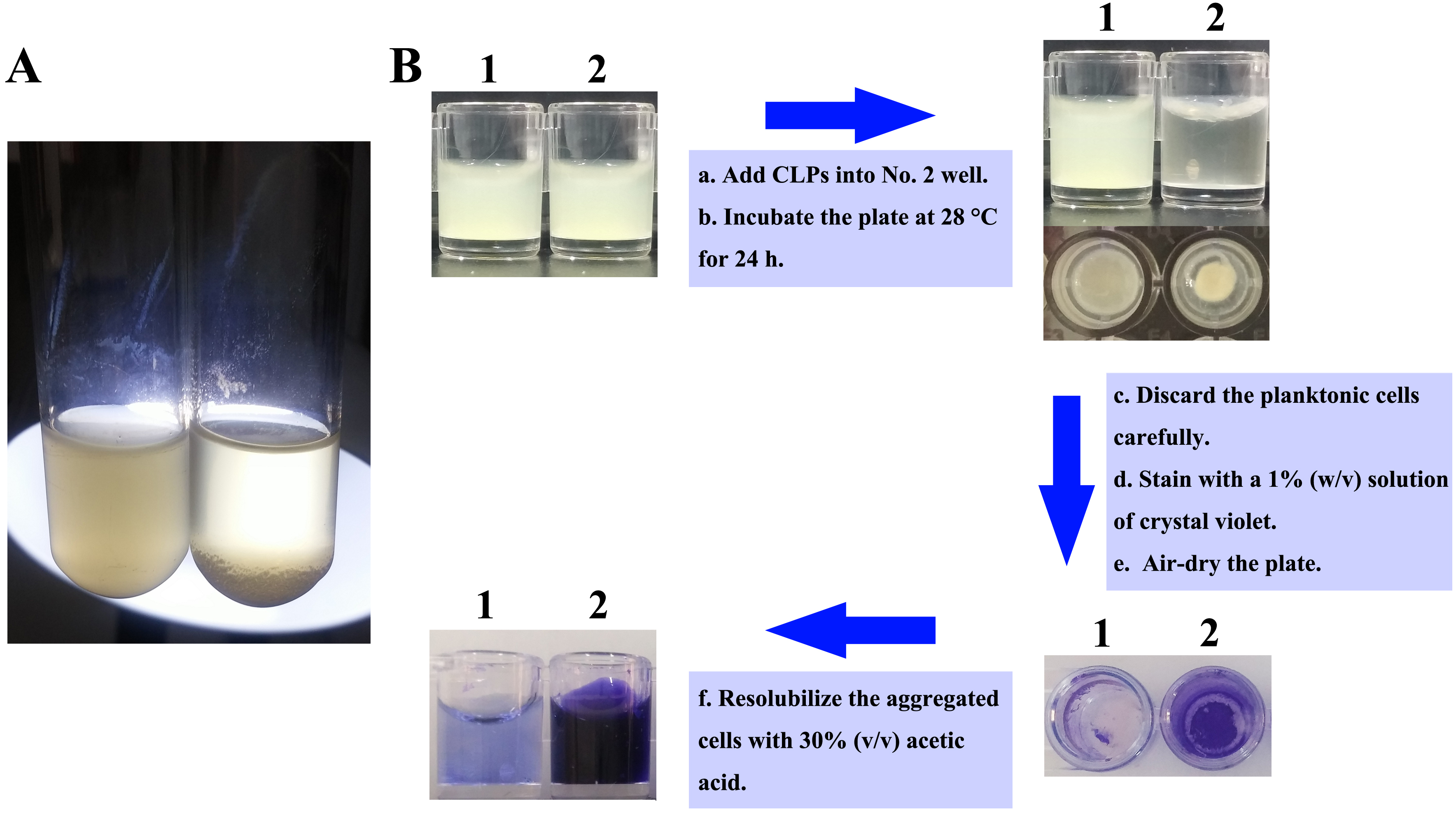
Figure 1. Experimental design of V. alginolyticus 178 aggregation in culture tubes and 96-well plates. A. Aggregation of V. alginolyticus 178 was observed in culture tubes after CLPs treatment. B. Schematic representation of the steps in aggregation assay using 96-well plates (No. 1 well contains untreated V. alginolyticus 178; No. 2 well contains CLPs treated V. alginolyticus 178).
- After overnight culture, dilute the cell suspension of V. alginolyticus 178 at 1:100 with saline LB broth.
- Aggregation assay of V. alginolyticus 178 in 96-well microtiter plate
- Add 200 μl diluted bacterial suspension into each well of the flat bottom 96-well microtiter plate.
- In experiment group, add 6 μl CLPs (10 mg/ml) with a final concentration of 300 μg/ml in each well, and homogenize with bacterial suspension.
- Wells containing V. alginolyticus 178 cell suspension without treatment, or with DMSO treatment, are employed as controls.
- Five replicate wells for each treatment, and incubate the plate at 28 °C for 24 h.
- After incubation, discard the planktonic cells carefully with a pipettor and wash the aggregated cells in each well three times with sterile saline.
- Fix aggregated cells with 200 μl of methanol (99% purity) per well statically for 15 min, and empty the plates using a pipettor and leave to dry.
- Then stain the contents of the wells with 200 μl of a 1% (w/v) crystal violet (see Recipes) solution for 10 min at room temperature.
- Rinse out crystal violet with a pipettor; air-dry the plates after wash with sterile deionized water, and resolubilize the dye bound to the aggregated cells in 200 μl of 30% (v/v) acetic acid (see Recipes) solution.
- Measure the absorbance of each well in a SYNERGY-H1 microplate reader (BioTek, USA) at 595 nm using 30% acetic acid as the blank (Figure 1B).
- Add 200 μl diluted bacterial suspension into each well of the flat bottom 96-well microtiter plate.
- Quantitative assay of CLPs’ activity
- Add 200 μl diluted bacterial suspension into each well of the flat bottom 96-well microtiter plate.
- Add 6 μl CLPs at different concentration (25, 50, 100, 200, 300 μg/ml) into different wells, respectively.
- Wells containing V. alginolyticus 178 cell suspension without treatment, or with DMSO treatment, are employed as controls.
- Operate the other procedures as described above in Steps C4-C9 (Figure 2).
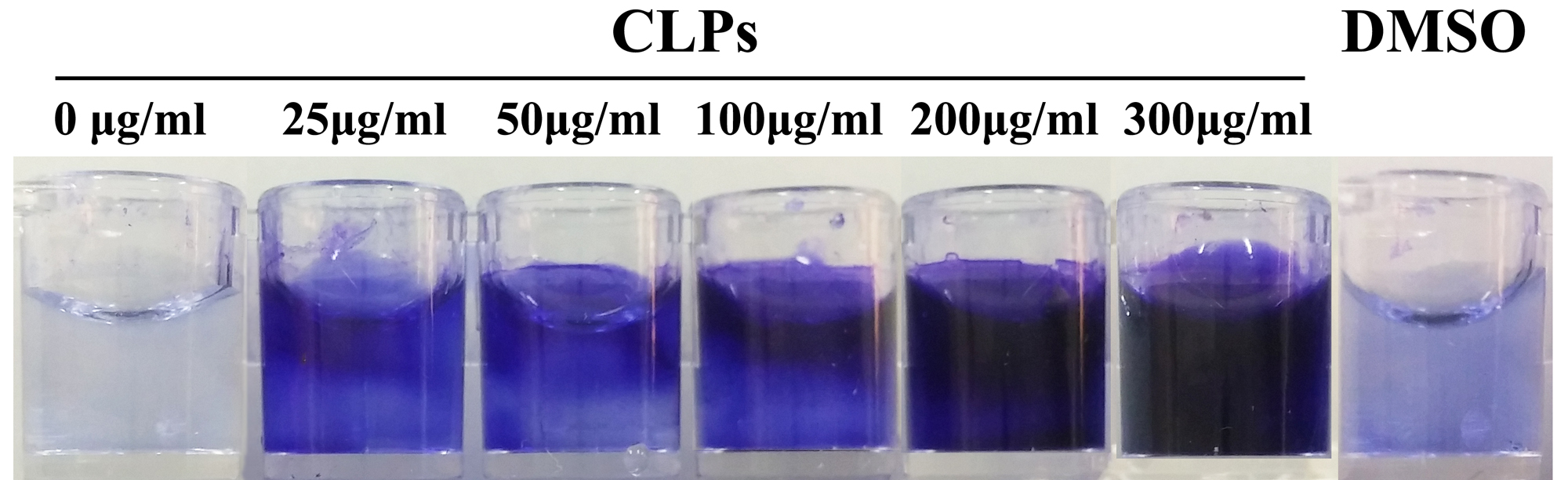
Figure 2. Quantitative cell aggregation assay at different concentrations of CLPs
- Add 200 μl diluted bacterial suspension into each well of the flat bottom 96-well microtiter plate.
- Scanning electron microscope observation
- Dilute overnight cultured cell of V. alginolyticus 178 at 1:100 into 5 ml fresh modified 2216E medium (see Recipes) and culture for another 3 h to OD600 0.2-0.3 with shaking at 170 rpm.
- Add the cell suspension with 50 μl 50 or 100 μg/ml CLPs or DMSO for additional 3 h.
- Centrifuge the cells at 1,400 x g for 4 min at room temperature, and resuspend the cells with a sterilized saline solution.
- Drop the resuspended bacterial cells on sterilized coverslips (enveloped by 1% gelation solution, see Recipes), respectively.
- Dry coverslips at room temperature.
- Fix samples with a 5% glutaraldehyde solution (see Recipes) for 1 h.
- Wash with sterile saline solution (see Recipes).
- Dehydrate samples in a successively graded ethanol series (50%, 60%, 70%, 80%, 90% and 100% ethanol). Incubate the samples for 10 min in each grade ethanol solution. After the final incubation in 100% ethanol, the samples are ready for observation under scanning electron microscope.
Note: You can stop when the samples are placed in 80% ethanol.
- Observe the samples under a scanning electron microscope (Figure 3).
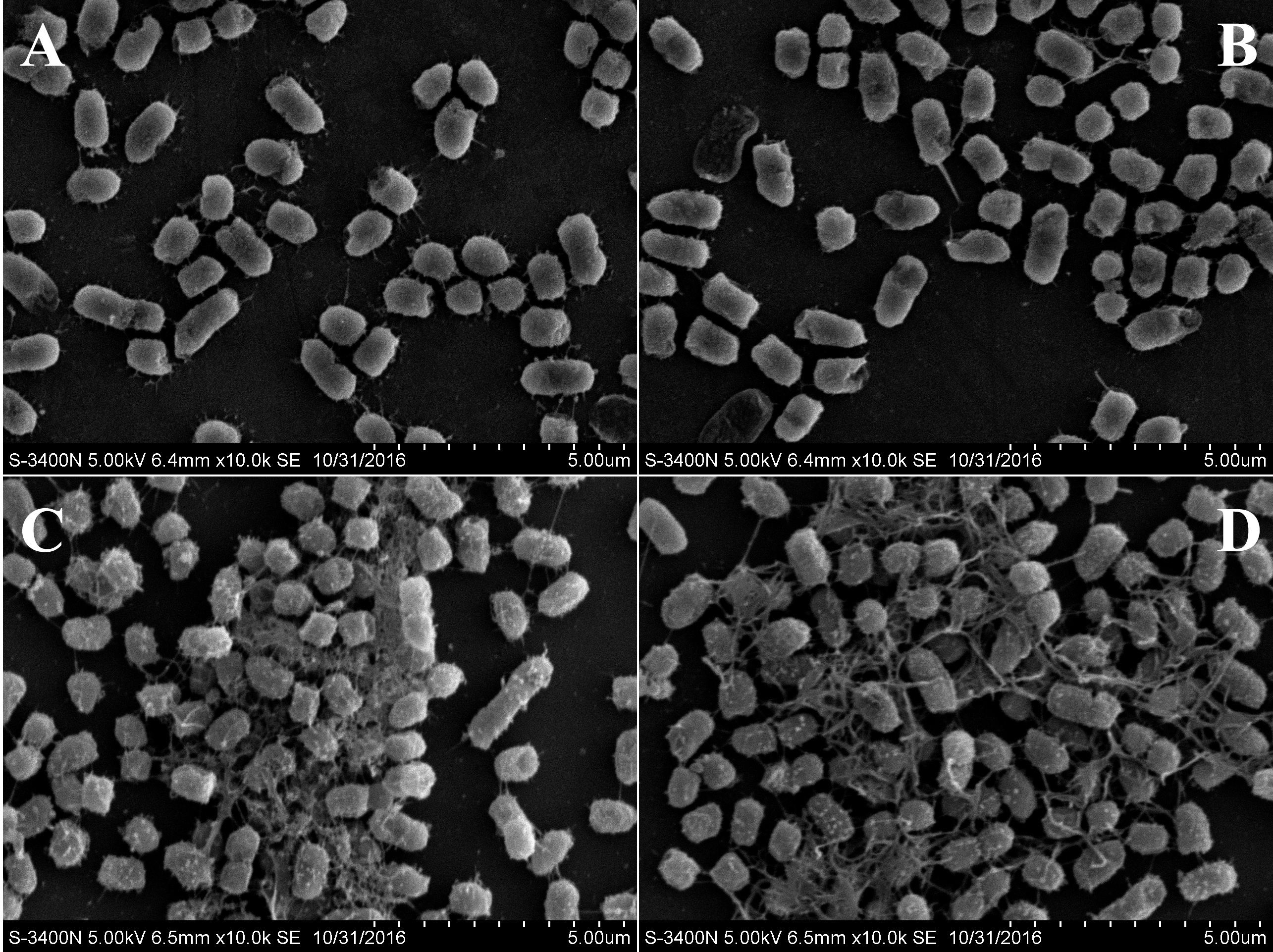
Figure 3. SEM images of V. alginolyticus 178 cells following treatment with CLPs. V. alginolyticus 178 cell morphology without any treatment (A), with DMSO treatment (B), with 50 μg/ml CLPs (C), and with 100 μg/ml CLP treatment (D).
- Dilute overnight cultured cell of V. alginolyticus 178 at 1:100 into 5 ml fresh modified 2216E medium (see Recipes) and culture for another 3 h to OD600 0.2-0.3 with shaking at 170 rpm.
- Transmission electron microscope observation
- Perform the procedures as described above in Steps E1-E3.
- Fix samples with a 5% glutaraldehyde solution for 1 h.
- Drop samples on copper grids, and dried at room temperature.
- Observe the samples under a transmission electron microscope (Figure 4).
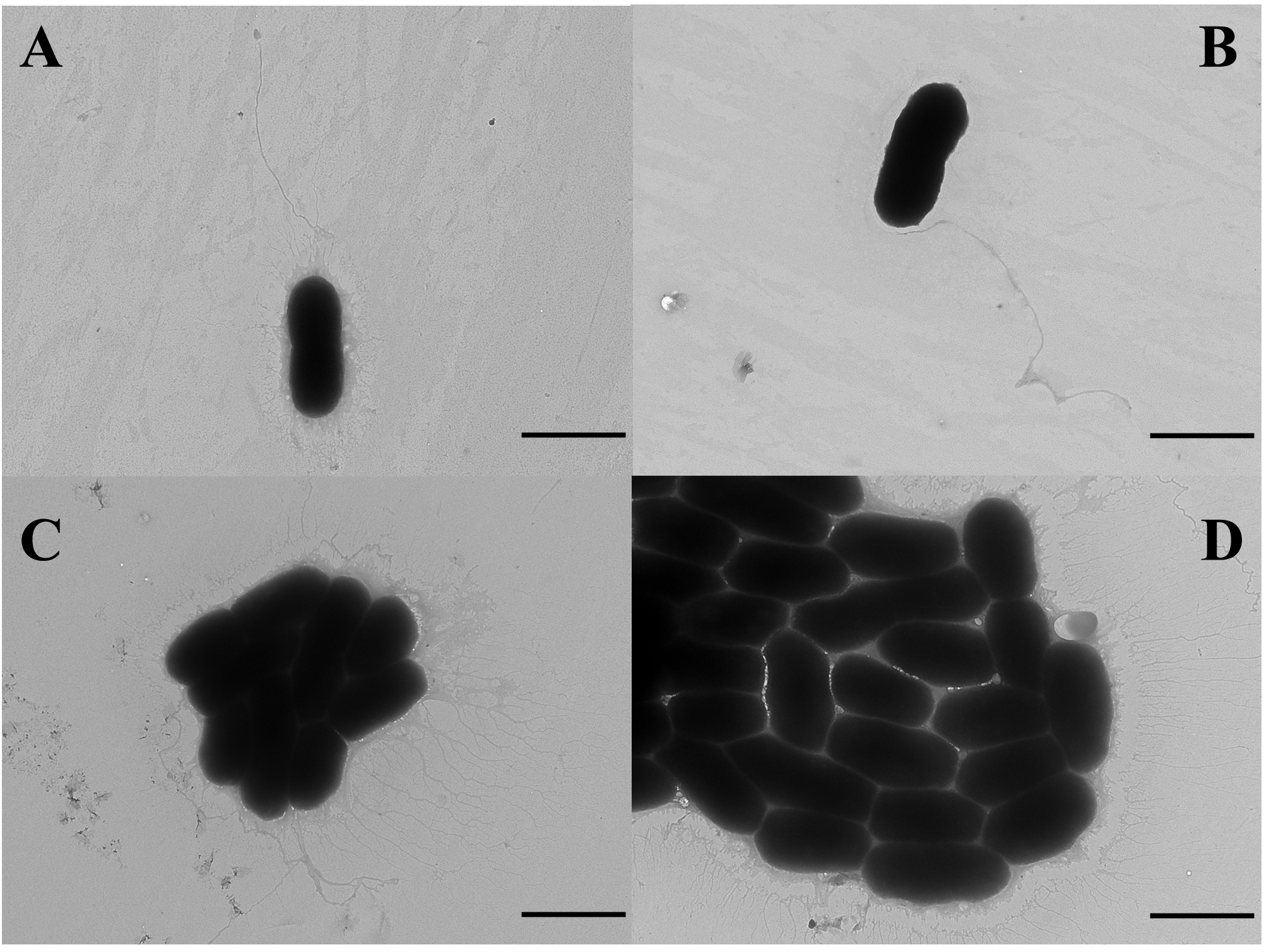
Figure 4. TEM images of V. alginolyticus 178 cells following treatment with CLPs. V. alginolyticus 178 cell morphology without any treatment (A), with DMSO treatment (B), with 50 μg/ml CLPs (C), and with 100 μg/ml CLP treatment (D). The scale bars are 2 μm.
- Perform the procedures as described above in Steps E1-E3.
- The spectrum of action of the CLPs
- After overnight culture, dilute the cell suspension of V. anguillarum, V. splendidus, V. vulnificus, Pseudomonas aeruginosa, P. stutzeri, Staphylococcus aureus, Bacillus sp. 176 and B. subtilis at 1:100 with saline LB broth or LB broth, respectively.
- Operate the other procedures as described above in Steps C2-C9 (Figure 5).
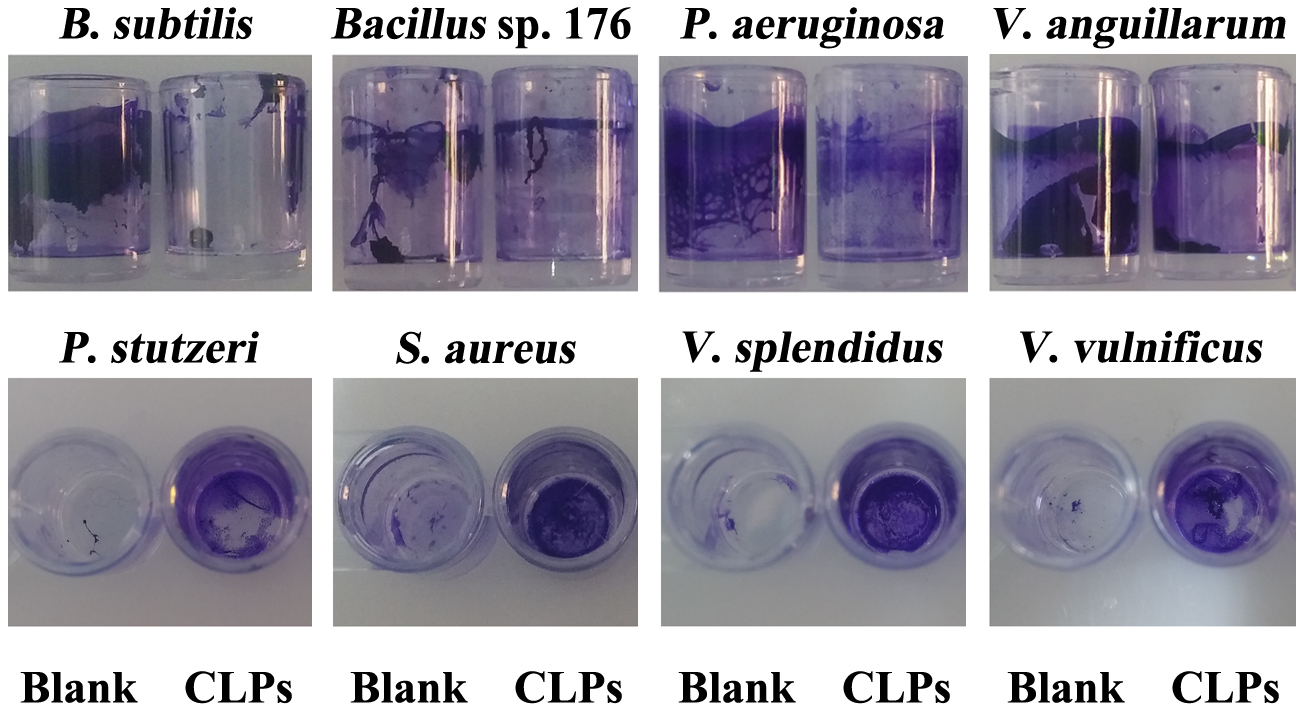
Figure 5. Action spectrum assays of CLPs. Blank: untreated strains; CLPs: CLPs treated strains.
- After overnight culture, dilute the cell suspension of V. anguillarum, V. splendidus, V. vulnificus, Pseudomonas aeruginosa, P. stutzeri, Staphylococcus aureus, Bacillus sp. 176 and B. subtilis at 1:100 with saline LB broth or LB broth, respectively.
Data analysis
All data were analyzed by the Statistical Package for Social Sciences (SPSS) 18.0 software. The statistically significant differences among groups were calculated using one-way analysis of variance (one-way ANOVA) followed by a post hoc multiple-comparisons (Tukey’s) test.
Notes
- To avoid other bacteria contamination, microbial operations must be carried out under aseptic conditions.
- All bacterial mediums and reagent solutions are sure to be prepared freshly and sterilized by autoclaving.
- It is necessary to homogenize the cell suspension with CLPs before incubation.
- Notice that the operation in discarding the planktonic cells and washing the aggregated cells with pipettor must be careful and gentle.
Recipes
- Saline LB broth (1 L)
10 g peptone
5 g yeast extract
1 L filtered seawater
pH 7.4-7.6
Sterilize by autoclaving at 121 °C for 20 min, and store at 4 °C
- LB broth (1 L)
10 g peptone
10 g NaCl
5 g yeast extract
1 L deionized water
pH 7.4-7.6
Sterilize by autoclaving at 121 °C for 20 min, and store at 4 °C
- 1% (w/v) solution of crystal violet (100 ml)
1 g crystal violet
100 ml sterile water
- 30% (v/v) acetic acid (100 ml)
30 ml glacial acetic acid
70 ml sterile water
- Modified 2216E broth (1 L)
5 g tryptone
1 g yeast extract
1 L filtered seawater
pH 7.4-7.6
Sterilize by autoclaving at 121 °C for 20 min, and store at 4 °C
- 1% (w/v) gelatin solution (10 ml)
0.1 g gelatin
10 ml deionized water
Sterilize by autoclaving at 121 °C for 20 min, and store at 4 °C
- 5% glutaraldehyde solution (10 ml)
2 ml 25% glutaraldehyde solution
8 ml sterile water
- Sterile saline solution (100 ml)
0.85 g NaCl
100 ml deionized water
Sterilize by autoclaving at 121 °C for 20 min, and store at 4 °C
- 10 mg/ml CLPs (1 ml)
10 mg CLPs
1 ml DMSO
Acknowledgments
This work was supported by Natural science outstanding youth fund of Shandong Province (JQ201607), Tai Shan Scholar Foundation of Shandong Province, AoShan Talents Program supported by Qingdao National Laboratory for Marine Science and Technology (No. 2015ASTP), ‘100-Talent Project’ of the Chinese Academy of Sciences to Chaomin Sun, and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA11030201) to Dechao Zhang. We also really appreciate Professor Nasrin Samadi and Dina Dalili, from Tehran University of Medical Sciences, for their method in study ‘Isolation and structural characterization of Coryxin, a novel cyclic lipopeptide from Corynebacterium xerosis NS5 having emulsifying and anti-biofilm activity’. And the authors declare that they have no competing interests.
References
- Arima, K., Kakinuma, A. and Tamura, G. (1968). Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun 31(3): 488-494.
- Cameotra, S. S. and Makkar, R. S. (2004). Recent applications of biosurfactants as biological and immunological molecules. Curr Opin Microbiol 7(3): 262-266.
- Dalili, D., Amini, M., Faramarzi, M. A., Fazeli, M. R., Khoshayand, M. R. and Samadi, N. (2015). Isolation and structural characterization of Coryxin, a novel cyclic lipopeptide from Corynebacterium xerosis NS5 having emulsifying and anti-biofilm activity. Colloids Surf B Biointerfaces 135: 425-432.
- Das, P., Mukherjee, S. and Sen, R. (2008). Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J Appl Microbiol 104(6): 1675-1684.
- Nam, H. S., Yang, H. J., Oh, B. J., Anderson, A. J. and Kim, Y. C. (2016). Biological control potential of Bacillus amyloliquefaciens KB3 isolated from the feces of Allomyrina dichotoma larvae. Plant Pathol J 32(3): 273-280.
- Naruse, N., Tenmyo, O., Kobaru, S., Kamei, H., Miyaki, T., Konishi, M. and Oki, T. (1990). Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J Antibiot (Tokyo) 43(3): 267-280.
- Rodrigues, L., Banat, I. M., Teixeira, J. and Oliveira, R. (2006). Biosurfactants: potential applications in medicine. J Antimicrob Chemother 57(4): 609-618.
- Roongsawang, N., Thaniyavarn, J., Thaniyavarn, S., Kameyama, T., Haruki, M., Imanaka, T., Morikawa, M. and Kanaya, S. (2002). Isolation and characterization of a halotolerant Bacillus subtilis BBK-1 which produces three kinds of lipopeptides: bacillomycin L, plipastatin, and surfactin. Extremophiles 6(6): 499-506.
- Sajitha, K. L., Dev, S. A. and Maria Florence, E. J. (2016). Identification and characterization of lipopeptides from Bacillus subtilis B1 against sapstain fungus of rubberwood through MALDI-TOF-MS and RT-PCR. Curr Microbiol 73(1): 46-53.
- Singh, P. and Cameotra, S. S. (2004). Potential applications of microbial surfactants in biomedical sciences. Trends Biotechnol 22(3): 142-146.
- Steller, S. and Vater, J. (2000). Purification of the fengycin synthetase multienzyme system from Bacillus subtilis b213. J Chromatogr B Biomed Sci Appl 737(1-2): 267-275.
- Vater, J., Kablitz, B., Wilde, C., Franke, P., Mehta, N. and Cameotra, S. S. (2002). Matrix-assisted laser desorption ionization--time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl Environ Microbiol 68(12): 6210-6219.
- Yakimov, M. M., Timmis, K. N., Wray, V. and Fredrickson, H. L. (1995). Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl Environ Microbiol 61(5): 1706-1713.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xiu, P., Liu, R., Zhang, D. and Sun, C. (2018). Bacterial Aggregation Assay in the Presence of Cyclic Lipopeptides. Bio-protocol 8(1): e2686. DOI: 10.21769/BioProtoc.2686.
Category
Microbiology > Antimicrobial assay > Antibacterial assay
Microbiology > Microbial biofilm > Biofilm culture
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









