- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fatty Acid Content and Composition of Triacylglycerols of Chlorella kessleri
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2676 Views: 9380
Reviewed by: Maria SinetovaRoman A. SidorovAgnieszka Zienkiewicz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sorghum bicolor Extracellular Vesicle Isolation, Labeling, and Correlative Light and Electron Microscopy
Deji Adekanye [...] Jeffrey L. Caplan
Oct 5, 2024 2107 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1718 Views

PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
Louise Fougère [...] Yohann Boutté
Sep 5, 2025 1334 Views
Abstract
Triacylglycerols (TAGs) are esters formed from one glycerol and three fatty acids. TAGs are induced to accumulate in algal cells under environmental stress conditions including nutrient-limitation, hyperosmosis, and low temperature, for the storage of metabolic energy and carbon, and also for the consumption of excess energy (e.g., Hirai et al., 2016; Hayashi et al., 2017). Beside their physiological significance, the commercial utilization of algal TAG has been expected for the production of biodiesel, the methyl esters of fatty acids, from the aspect of carbon-neutral conception. The amounts of TAGs can be determined through quantitative measurement of their constituent fatty acids. This protocol consists of the following three parts: the first is the extraction of total lipids from algal cells with the use of organic solvents, chloroform and methanol, according to the method of Bligh and Dyer (1959), the second is the separation of TAG from the other lipid classes by thin-layer chromatography (TLC), and the third is the production of methyl-esterified derivatives of their constitutive fatty acids and subsequent quantitation of them by capillary gas-liquid chromatography (GLC). This protocol adapted from Sato and Tsuzuki (2011) is used for TAG analysis in a green alga, Chlorella kessleri.
Keywords: Chlorella kessleriBackground
Several methods have been used for determination of the fatty acid content of TAG. Simple and convenient protocols, e.g., include conversion of TAG to glycerol on treatment with a lipase, and subsequent measurement of the glycerol content through enzymatic generation of a product that reacts with a color- or fluorescence-generating probe (McGowan et al., 1983; Mendez et al., 1986). However, this enzymatic reaction based quantitation of TAG, inevitably, gives no information about the composition of constituent fatty acids. Meanwhile, HPLC provides information on TAG molecular species through their separation based on the numbers of carbon atoms and double bonds of constituent fatty acids, and enables their respective quantitation when combined with tandem mass spectrometer like in LC-MS/MS (Mu et al., 2000; Dorschel, 2002; MacDougall et al., 2011). The LC-MS/MS instrument, however, is very expensive. In this context, TLC/GLC based protocol for the measurement of the fatty acid content of TAG is introduced here, in view of the requirement of less expensive equipment than LC-MS/MS and definite information that can be obtained on quality and quantity of the constituent fatty acids.
Materials and Reagents
- 50 ml polypropylene centrifuge tubes with conical bottom (Corning, Falcon®, catalog number: 352070 )
- 50 ml glass screw cap centrifuge tubes with PTFE lined phenolic caps (AGC Techno Glass, catalog number: 8422CTF50 )
- 9 inch glass Pasteur pipettes (AGC Techno Glass, catalog number: IK-PAS-9P )
- TLC silica gel 60 glass plates 20 x 20 cm (Merck, catalog number: 105721 )
- Chromatography filter paper 1CHR 200 x 200 mm (GE Healthcare, catalog number: 3001-861 )
- Glass microcapillary pipette (Sigma-Aldrich, catalog number: Z543292 )
- 100 µl microsyringe (1710 RN, Hamilton, catalog number: 81030 ) with 22s/51/2 needle (Hamilton, catalog number: 7758-03 )
- 14 ml glass screw cap test tubes with PTFE lined phenolic caps (AGC Techno Glass, catalog number: TST-SCR16-125 )
- Inserts for large opening vials volume 0.15 ml (Sigma-Aldrich, catalog number: 24719 )
- 2 ml large opening vials with open-top screw cap (Sigma-Aldrich, catalog number: 29116-U )
- ULBON HR-Thermon-3000B GLC capillary column I.D. 0.25 x 25 m (Shinwa Chemical Industries)
- Chlorella kessleri 11 h, which corresponds to Parachlorella kessleri of NIES collection (http://mcc.nies.go.jp/) (National Institute for Environmental Studies, catalog number: NIES-2160 )
- Potassium chloride (KCl) (Wako Pure Chemical Industries, catalog number: 163-03545 )
- Butylated hydroxytoluene (BHT) (Wako Pure Chemical Industries, catalog number: 029-07392 )
- Methanol (CH3OH) (Wako Pure Chemical Industries, catalog number: 138-06473 )
- Chloroform (CHCl3) (Wako Pure Chemical Industries, catalog number: 033-08631 )
- Primuline (C21H15N3O3S3) (Tokyo Chemical Industry, catalog number: P0603 )
- Acetone (CH3COCH3) (Wako Pure Chemical Industries, catalog number: 016-00346 )
- Tripalmitin (C51H98O6) as TAG standard (Wako Pure Chemical Industries, catalog number: 200-03002 )
- N2 gas
- Hydrogen chloride-methanol reagent (5-10%) (Tokyo Chemical Industry, catalog number: X0041 )
- n-Hexane (C6H14) (Wako Pure Chemical Industries, catalog number: 084-03421 )
- Supelco 37 component FAME mix (Sigma-Aldrich, catalog number: 47885-U )
- Gamborg’s B5 medium salt mixture (Nihon Pharmaceutical, catalog number: 399-00621 ; Gamborg et al., 1968)
- Sorbitol (C6H14O6) (Wako Pure Chemical Industries, catalog number: 198-03755 )
- Arachidic acid (C20H40O2) (Tokyo Chemical Industry, catalog number: E0006 )
- 1/4 GB medium with or without 0.6 M sorbitol (see Recipes)
- Arachidic acid solution as an IS (see Recipes)
Equipment
- High-pressure steam sterilizer (TOMY SEIKO, model: LBS-245 )
- Tabletop centrifuge (Kubota, model: 5220 ) equipped with ST-720M swing rotor (16 x 50 ml)
- Vortex mixer (Scientific Industries, model: Vortex-Genie 2 )
- Rotary evaporator (Tokyo Rikakikai, model: N-1110V-W ) equipped with screw cap tube adaptor
- UV transilluminator (UVP, model: LM-20 )
- Fume hood (Yamato Scientific, model: KFS )
- Forced air flow oven (Tokyo Rikakikai, model: WFO-451SD )
- Spectrophotometer (Beckman Coulter, model: DU 640 )
- Double trough TLC chamber for 20 x 20 cm plates (Camag, catalog number: 022.5256 )
- Capillary gas-liquid chromatograph (Shimadzu, model: GC-2025 ) equipped with split/splitless injector, frame ion detector (FID) and autoinjector
- Chromatopac integrator (Shimadzu, model: C-R7A plus )
- Glass chromatographic reagent atomizer (Corning, PYREX®, catalog number: 2153-125 )
Procedure
- Total lipid extraction
- Culture C. kessleri cells at 30 °C in 100 ml of 1/4 GB (see Recipes) without sorbitol, under illumination (30 W m-2) and aeration for 48-72 h, to the late exponential phase of growth (the values of optical density at 730 nm [OD730] within ca. 0.4 to 0.6).
- Harvest cells through centrifugation in 2 x 50 ml tubes at 1,500 x g for 15 min at room temperature (tabletop centrifuge). Discard supernatant by decantation and resuspend cells in 20 ml of 1/4 GB with 0.6 M sorbitol for the induction of TAG accumulation (Hirai et al., 2016).
- Harvest cells through centrifugation at 1,500 x g for 15 min. Discard supernatant through decantation and resuspend cells in 20 ml of 1/4 GB with 0.6 M sorbitol. Repeat this centrifugation-resuspension step twice, finally with the OD730 value of cell culture adjusted to 0.3 in 50 ml of 1/4 GB with 0.6 M sorbitol. Culture cells for three days under the same growth conditions.
- Harvest cells through centrifugation at 1,500 x g for 15 min at room temperature. Discard supernatant through decantation and resuspend cells in 2 ml of 0.1 M KCl.
- Transfer cell suspension into a glass screw cap centrifuge tubes (50 ml vol.) with PTFE lined phenolic caps. Add 6.0 ml of methanol and 30 µl of 1.0% (w/v) BHT in methanol as an antioxidant agent to the sample, and agitate it by a vortex mixer for 30 sec at the maximum speed to destabilize cellular membrane systems.
- Add 3.0 ml of chloroform to the sample. Agitate it by a vortex mixer for 30 sec at the maximum speed and stand it for 10 min to extract lipids from the cells.
- Add 3.0 ml of chloroform once again and agitate the sample for intensive extraction of lipids by a vortex mixer for 30 sec at the maximum speed.
- Add 3.0 ml of distilled water and vortex the sample for 30 sec for emulsification. Centrifuge the mixture at 1,000 x g for 5 min at room temperature for its separation into three phases, i.e., the upper phase of H2O and methanol, the medium phase of cell debris, and the lower phase of chloroform. There is no cell debris at the bottom of the tube.
- Transfer the lower phase including total lipids with a Pasteur pipette into a glass screw cap centrifuge tubes (50 ml vol.) with PTFE lined phenolic caps. Be careful not to pick up the cell debris or the upper phase. The remaining lower phase can be recovered by the subsequent procedure.
- Add 3.0 ml of chloroform to the remaining upper and medium phases, and agitate the mixture for 30 sec with a vortex mixer. Centrifuge it at 1,000 x g for 15 min at room temperature.
- Recover the lower phase to the glass tube at Step A9.
- Repeat Steps A10 to A11 twice until lipids are fully recovered. Full recovery of lipids can be confirmed through observation of bleaching of the cell debris, which results from extraction of green chlorophylls into the lower phase.
Note: Do not mechanically disrupt C. kessleri cells before lipid extraction, since the mechanical disruption would not increase the TAG recovery, but would decrease the TAG content with appearance of free fatty acids, owing probably to the action of endogenous lipases. - Evaporate the solvent completely with a rotary evaporator, if necessary, with the addition of methanol or chloroform/methanol (2:1, v/v) for removal of residual H2O through azeotropy. Weigh dry residue and dissolve lipids in ca. 200 µl of chloroform/methanol (2:1, v/v), and transfer this total lipid fraction into a glass vial to be stored at -20 °C until use. The value of dry residue weight will be helpful for optimization of loading of total lipids on TLC plate for better separation of TAG.
- Culture C. kessleri cells at 30 °C in 100 ml of 1/4 GB (see Recipes) without sorbitol, under illumination (30 W m-2) and aeration for 48-72 h, to the late exponential phase of growth (the values of optical density at 730 nm [OD730] within ca. 0.4 to 0.6).
- Separation of TAG from the other lipid classes by TLC
- Set a TLC chamber for the separation of TAG by placing a piece of filter paper along its wall, and then by pouring a developing solvent of hexane/diethylether/acetate (70:30:1, v/v) up to ca. 1 cm in depth. Close the lid for vapor saturation, which is promoted by the filter paper that has been soaked in the solvent. Stand the chamber for an hour or more to saturate it with the solvent vapor prior to the start of development. In order to ensure repeatability, the solvent is prepared freshly for each analysis.
- Heat a TLC silica gel 60 plate at 120 °C for a couple of hours in an oven to activate the silica gel just before development. Don’t touch the plate with bare hands during this procedure.
- Draw a horizontal line that is about 2 cm away from the bottom side of a TLC plate with a carbon pencil, and outline a horizontally long rectangle (ca. 2 x 3 cm) on the horizontal line of the plate. Lines have to be gently drawn to avoid scraping off silica gel. Don’t use an ink pen, otherwise, the lines would disappear.
Note: Depending on the number of samples, the TLC plate may be cut into two pieces (10 cm wide ones) before use. - Apply 45% of the stored total lipid fraction to the rectangle spot with a glass microcapillary pipette or 100 µl microsyringe with 22s/51/2 needle, and air-dry it. Repeat this step several times until this fraction is completely transferred to this spot.
- Apply 5-25 µl of standard TAG stock solution (1 mg/ml in chloroform, w/v) on the horizontal line next to the spot of the analyte samples.
- Place the TLC plate in the chamber, cover the chamber with its lid, and allow the plate to develop until the solvent goes up to about 2 cm below the top side of the plate.
- Remove the plate from the chamber and air-dry the plate.
- Spray the plate with primuline (0.01% in 80% acetone, w/v) in a fume hood, and illuminate it under 365 nm UV light to detect lipid compounds as exhibiting a pale color (see Figure 1). Outline the TAG spot with a pencil, judged from the position of TAG standard.
Note: To prevent the TAG spot from diffusing, avoid spraying excessive detection reagent. - Scrap off the silica gel of the TAG spot by a flat edge of micro-spatula. Silica gel from TAG should be immediately treated for the production of fatty acid methyl esters (FAMEs, see below) to prevent the oxidation of its constituent fatty acids.
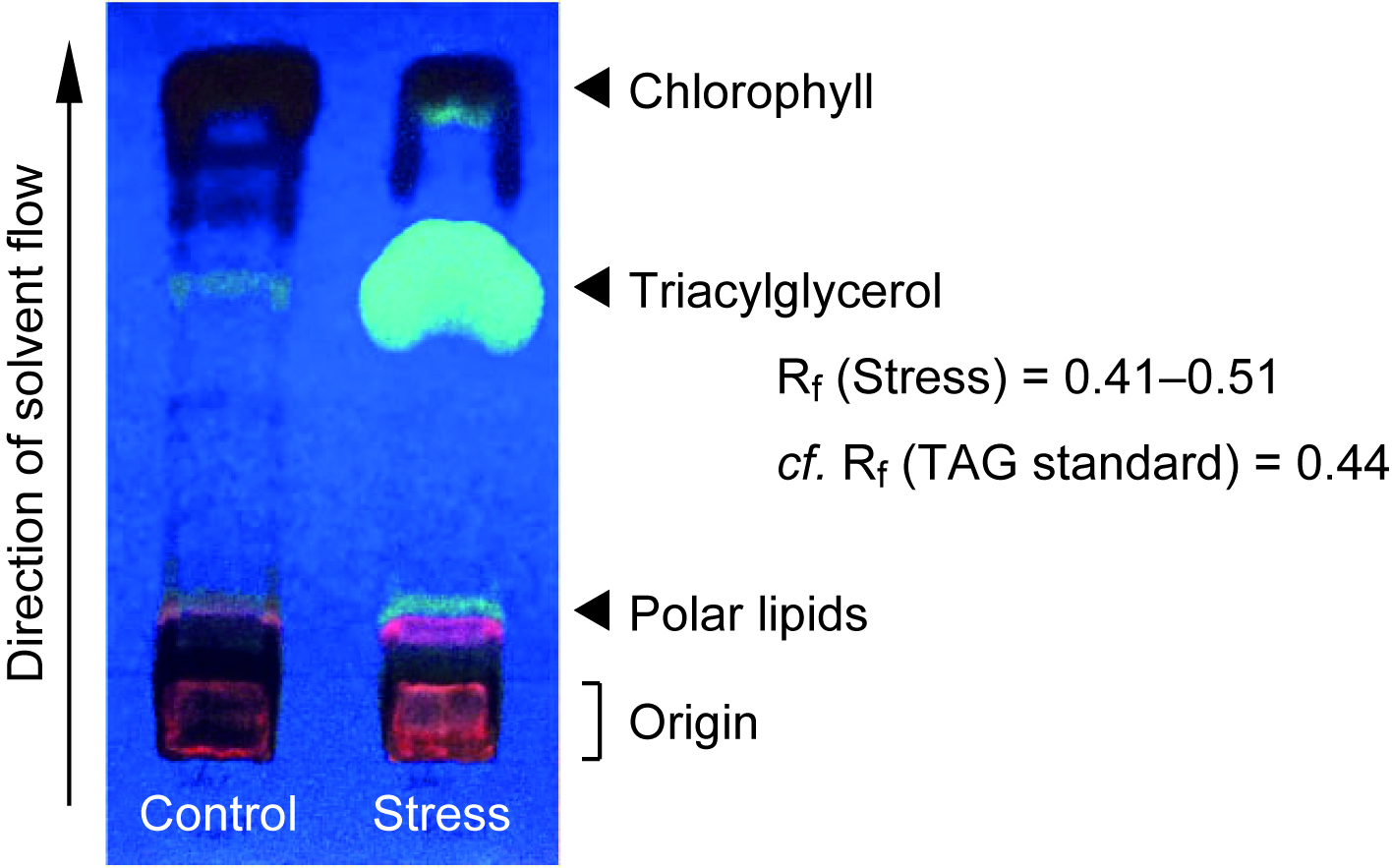
Figure 1. TLC separation of TAG in total lipid fraction in C. kessleri
- Set a TLC chamber for the separation of TAG by placing a piece of filter paper along its wall, and then by pouring a developing solvent of hexane/diethylether/acetate (70:30:1, v/v) up to ca. 1 cm in depth. Close the lid for vapor saturation, which is promoted by the filter paper that has been soaked in the solvent. Stand the chamber for an hour or more to saturate it with the solvent vapor prior to the start of development. In order to ensure repeatability, the solvent is prepared freshly for each analysis.
- Conversion of constituent fatty acids into fatty acid methyl esters
- Take 50 µl of arachidic acid solution as an internal standard (IS, see Recipes) into glass screw cap test tubes (14 ml vol.) with PTFE lined phenolic cap, and dry it up under the flow of N2 gas.
Note: Since Chlorella kessrelli 11 h has no endogenous C20 series fatty acids, the authors use arachidic acid (C20:0) as an IS. Nonadecanoic acid (C19:0) can also be used as an IS. - Put scraped silica gel of TAG and 5% of the total lipid fraction into the test tubes, respectively. The latter lipid solution has to be air-dried under the flow of N2 gas. The remainder of the total lipid fraction, i.e., 50% of the initial level, can be stored at -80 °C for later analysis.
- Add 2.0 ml of 5-10% (w/v) hydrogen chloride-methanol reagent to each tube, cap it tightly, and agitate it by a vortex mixer for 30 sec at the maximum speed.
- Heat each tube at 95 °C for 2 h for methyl-esterification of the constituent fatty acids of TAG or total lipids.
- Cool down the tubes to room temperature, and add 2 ml of n-hexane to it. Agitate the tubes by a vortex mixer for 30 sec at a maximum speed for the extraction of FAMEs.
- Stand the tubes for 1 min for the separation of its content into upper (n-hexane) and lower (methanol) phases.
- Transfer the upper phase into a glass screw cap centrifuge tube (50 ml vol.) with PTFE lined phenolic caps by a glass Pasteur pipette.
- Add 2.0 ml of n-hexane to the remaining lower phase and vortex it for 30 sec. Stand the tube for 1 min at a room temperature.
- Recover the upper phase to the glass tube at Step C7.
- Repeat Steps C8 to C9 additionally two times for intensive extraction of FAMEs.
- Evaporate FAMEs solution with a rotary evaporator, and dissolve the dried samples in a small volume (< 50 µl) of n-hexane. Transfer the concentrated FAMEs solution into a 0.15 ml insert in a 2 ml glass vial, which is then capped for storage at 4 °C until measurement.
- Analyze fatty acid composition of the sample with a capillary GLC (see Figure 2). GLC is operated according to manufacturer’s instructions. GLC operating conditions are shown in Table 1.
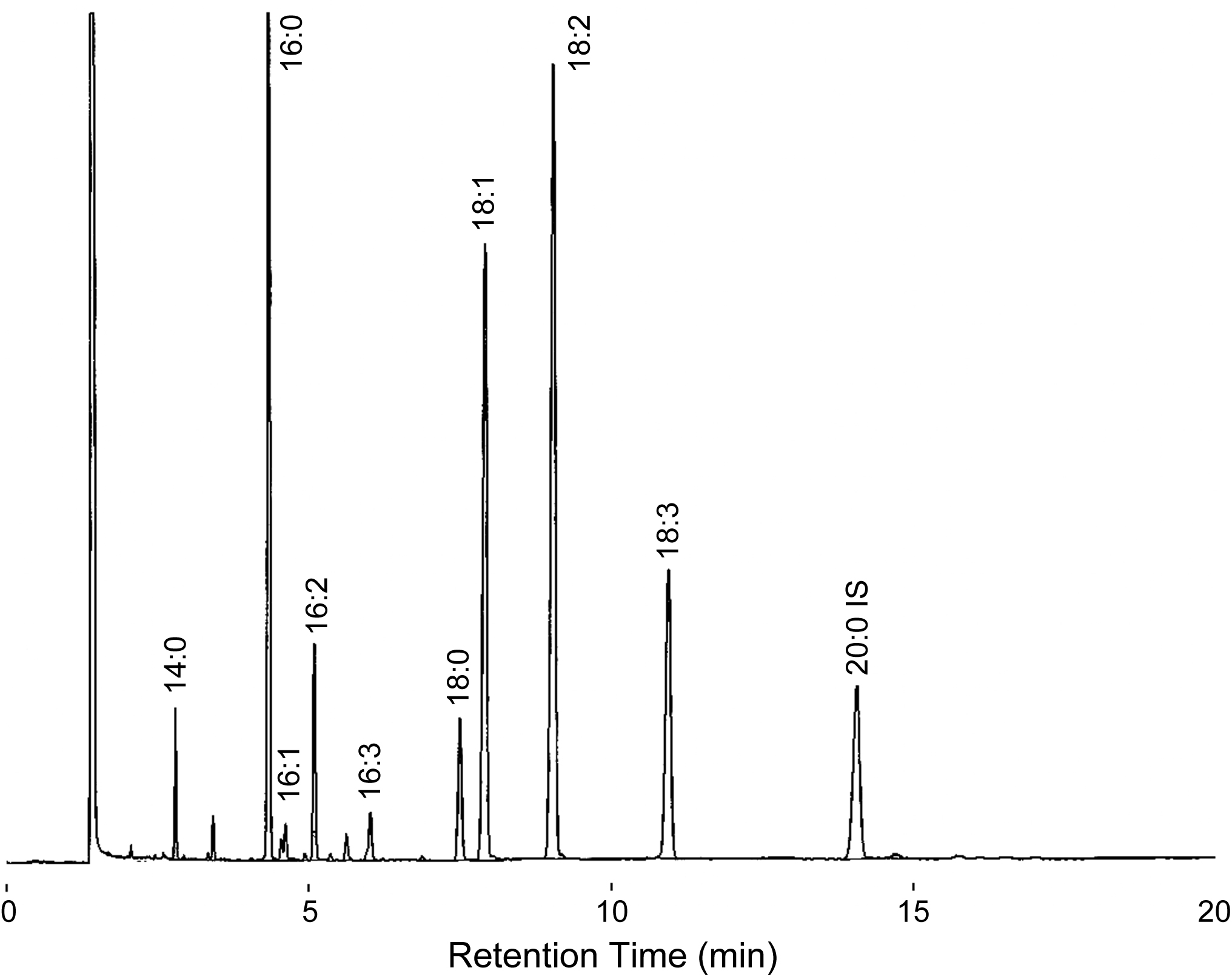
Figure 2. A gas chromatogram of FAMEs from TAG of C. kessleri
Table 1. GLC operating conditions for determination of fatty acid composition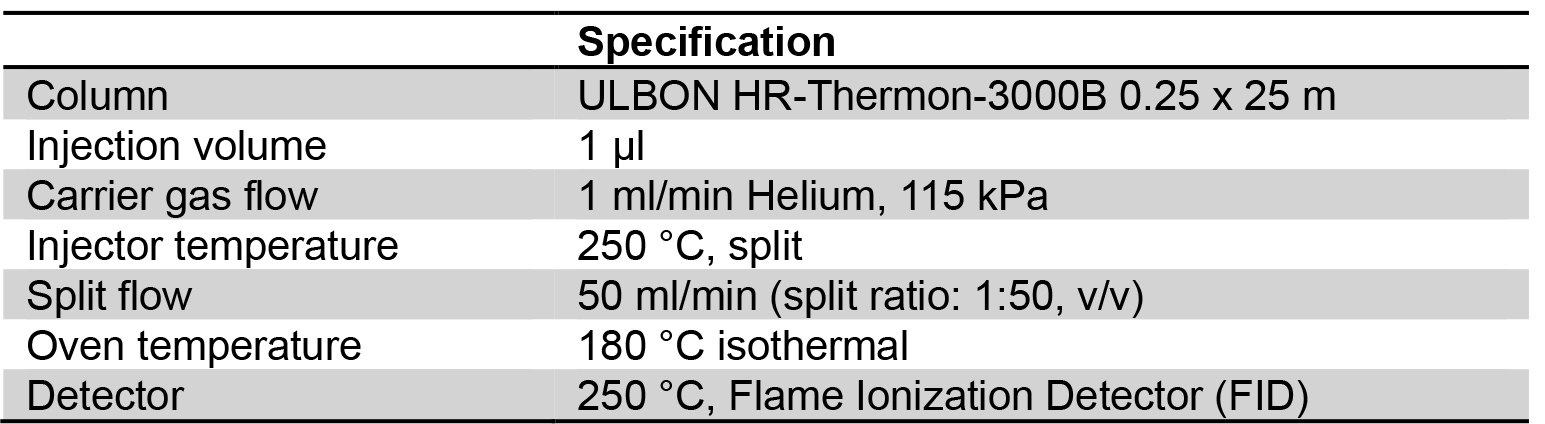
Note: FID detects ions that are generated through burning of carbon compounds (Holm, 1999).
- Take 50 µl of arachidic acid solution as an internal standard (IS, see Recipes) into glass screw cap test tubes (14 ml vol.) with PTFE lined phenolic cap, and dry it up under the flow of N2 gas.
Data analysis
- Identify and calculate the peak areas of respective FAMEs on chromatogram with an integrator according to manufacturer’s instructions. FAMEs identification is based on the retention times as compared with those of the standard FAME mixture (Supelco 37 component FAME Mix, 100 µg/ml solution diluted with n-hexane). Check that chromatogram is being integrated properly with the appropriate baseline being shown.
- Quantitate each molecular species of FAME by using following Formula [1] with arachidic acid (20:0) as an IS. The values, 160 and 312, indicate the content of 20:0 (nmol) taken at Step C1 and the molecular weight of 20:0, respectively. The molar content of total lipids can be expressed on the basis of fatty acids that are included (Formula [2]).
- The TAG content is estimated as 1/3 of the summed molar contents of its constituent fatty acids (Formula [3]). Be careful that obtained values correspond to those included in respective samples that are analyzed. Alternatively, the content of TAG relative to that of total lipids is estimated on the basis of fatty acids (Formula [4]). As shown here, multiplication of ‘summed content of FAME from TAG’ by 1/9 is necessary in accordance with the quantitative ratio of the total lipid fraction used for TAG analysis (45%) to that for total lipid analysis (5%). The quantitative results should be presented by mean values ± standard deviations from at least three experiments.
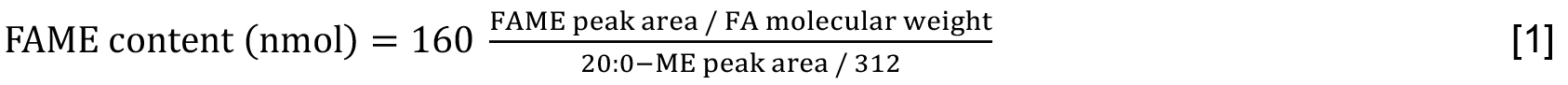
Note: The peak area of FAME is conveniently divided by the molecular weight of FA, but not by that of FAME, in view of full responses of FAME carbon atoms except its carbonyl one in FID (Ackman and Sipos, 1964).
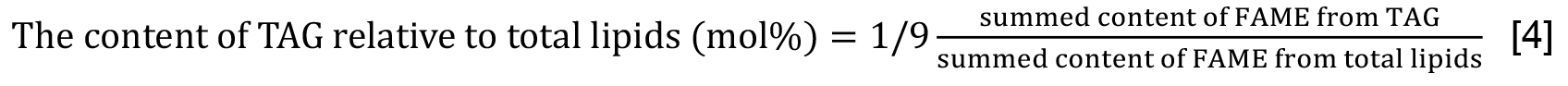
Recipes
- 1/4 GB medium with or without 0.6 M sorbitol
- Dissolve Gamborg’s B5 medium salt mixture (3.3 g/packet, Table 2) in distilled water for preparation of normal Gamborg’s B5 medium (GB; final volume, 1 L)
Table 2. Chemical composition of Gamborg’s B5 medium salt mixture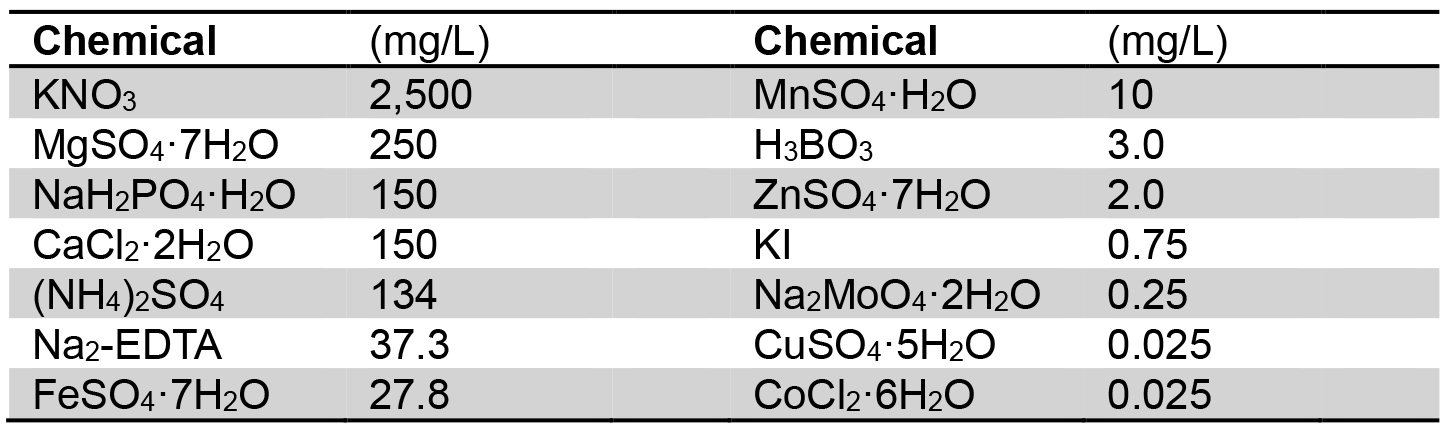
- Add 750 ml of distilled water to 250 ml of GB for preparation of 1/4 GB
- Otherwise, dissolve 109 g of sorbitol in 250 ml of GB with concomitant addition of distilled water for preparation of 0.6 M sorbitol containing 1/4 GB (final volume, 1 L)
- 1/4 GB and 0.6-M sorbitol containing 1/4 GB have to be subjected to autoclave sterilization (120 °C, 20 min)
- Dissolve Gamborg’s B5 medium salt mixture (3.3 g/packet, Table 2) in distilled water for preparation of normal Gamborg’s B5 medium (GB; final volume, 1 L)
- Arachidic acid solution as an IS
- Dissolve 10 mg of arachidonic acid in 10 ml 2-propanol
- Store the solution at 2-8 °C
- Dissolve 10 mg of arachidonic acid in 10 ml 2-propanol
Acknowledgments
This work was supported by Tokyo University of Pharmacy and Life Sciences. The authors have declared that no competing interests exist.
References
- , R.G. and Sipos, J. C. (1964). Application of specific response factors in the gas chromatographic analysis of methyl esters of fatty acids with flame ionization detectors. J Am Oil Chem Soc 41 (5): 377-378.
- Bligh, E. G. and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37 (8): 911-917.
- Dorschel, C. A. (2002). Characterization of the TAG of peanut oil by electrospray LC-MS-MS. J American Oil Chemists’ Society 79 (8): 749-753.
- Gamborg, O. L., Miller, R. A. and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50 (1): 151-158.
- Hayashi, T., Otaki, R., Hirai, K., Tsuzuki, M. and Sato, N. (2017). Optimization of seawater-based triacylglycerol accumulation in a freshwater green alga, Chlorella kessleri, through simultaneous imposition of lowered-temperature and enhanced-light intensity. Algal Res 28: 100-107.
- Hirai, K., Hayashi, T., Hasegawa, Y., Sato, A., Tsuzuki, M. and Sato, N. (2016). Hyperosmosis and its combination with nutrient-limitation are novel environmental stressors for induction of triacylglycerol accumulation in cells of Chlorella kessleri. Sci Rep 6: 25825.
- Holm T. (1999). Aspects of the mechanism of the flame ionization detector. J Chromatogr A 842 (1-2): 221-227.
- MacDougall, K. M., McNichol, J., McGinn P. J., O’Leary, S. J. B. and Melanson, J. E. (2011). Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography-high-resolution mass spectrometry. Anal Bioanal Chem 401 (8): 2609-2616.
- McGowan, M. W., Artiss, J. D., Strandbergh, D. R. and Zak, B. (1983). A peroxidase-coupled method for the colorimetric determination of serum triglycerides. Clin Chem 29 (3): 538-542.
- Mendez, A. J., Cabeza, C. and Hsia, S. L. (1986). A fluorometric method for the determination of triglycerides in nanomolar quantities. Anal Biochem 156 (2): 386-389.
- Mu, H., Sillen, H. and Hiy, C. E. (2000). Identification of diacylglycerols and triacylglycerols in a structured lipid sample by atmospheric pressure chemical ionization liquid chromatography/mass spectrometry. J American Oil Chemists’ Society 77(10): 1049-1060.
- Sato, N. and Tsuzuki, M. (2011). Isolation and identification of chloroplast lipids. Methods Mol Biol 684: 95-104.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Aoki, M. and Sato, N. (2018). Fatty Acid Content and Composition of Triacylglycerols of Chlorella kessleri. Bio-protocol 8(1): e2676. DOI: 10.21769/BioProtoc.2676.
Category
Plant Science > Plant biochemistry > Lipid
Microbiology > Microbial biochemistry > Lipid
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











