- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Infection of Soybean Plants with the Insect Bacterial Symbiont Burkholderia gladioli and Evaluation of Plant Fitness
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2663 Views: 7850
Reviewed by: Andrea PuharDaniel F. CaddellAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 404 Views
Abstract
To investigate the establishment and consequences of host-microbe interactions, it is important to develop controlled infection assays suitable for each system, as well as appropriate methods to evaluate successful infection and its associated effects. Here, we describe a procedure for bacterial inoculation of soybean plants, followed by the assessment of systemic infection and impact on plant fitness. Soybean (Glycine max) seedlings were mechanically wounded using a device that mimics insect herbivory and inoculated with known cell numbers of Burkholderia gladioli bacteria previously isolated from an insect host. The impact on the plants was evaluated by monitoring changes in height, time to flowering and chlorophyll content during plant development, and by quantifying seed production in comparison to plants inoculated with sterile water. The presence and proliferation of bacterial infection were examined in tissues from developed plants using quantitative PCR and fluorescence in situ hybridization (FISH).
Keywords: Bacterial plant infectionBackground
Microbes establish symbiotic associations with diverse eukaryotic organisms and can have profound effects on host fitness, ranging from beneficial to detrimental (Frank, 1997). In many cases, these associations are directly or indirectly influenced by interactions with additional organisms, like potential alternative hosts. As an example, there are numerous three-way interactions between plants, microbes and insects, in which microbial symbionts are transmitted between the different hosts and affect the physiology or ecology of the organisms involved (Frago et al., 2012; Gilbert et al., 2012). In phytophagous Lagriinae beetles, a symbiotic partnership has been established with bacteria from a plant pathogenic clade, Burkholderia gladioli, suggesting that this association evolved in the context of a tripartite interaction. Horizontal transmission of the symbionts from Lagria villosa beetles to soybean plants was previously demonstrated, indicating the possibility for dynamic transitions between hosts (Flórez et al., 2017). Furthermore, the symbionts are transmitted vertically, from mother to offspring on the surface of the beetle eggs (Stammer, 1929), where they inhibit the growth of pathogenic fungi (Flórez et al., 2017). In line with the dynamic nature of this symbiosis, at least three different strains of symbiotic B. gladioli are present in L. villosa, of which only one has been successfully cultured in vitro so far (Flórez and Kaltenpoth, 2017). This protocol has been developed to assess the ability of culturable B. gladioli bacteria to infect soybean plants, a common food source for L. villosa beetles, and evaluate the impact of infection on plant fitness.
Materials and Reagents
- Materials
- Plant pots 6 cm diameter (Volume 120 ml)
- Plant saucers Ø 10 cm and 16 cm (lower and upper diameter, respectively)
- Centrifuge tubes 50 ml (Corning, Falcon®, catalog number: 352070 )
- Thin wood sticks 30 cm (Schreiber-Online-Handel, Splittstäbe Naturweide, catalog number: 231979935696 )
- Razor blade (Schreiber, 11-0100)
- Reaction tubes 1.5 ml (SARSTEDT, catalog number: 72.690.001 )
- Petri dishes Ø 9 mm (Carl Roth, GosselinTM, catalog number: ALA5.1 )
- 1 ml pipette tips (SARSTEDT, catalog number: 70.762.010 )
- Polysine® glass slides (Thermo Fisher Scientific, Menzel-Gläser, catalog number: J2800AMNZ )
- Microscope cover slips 24 x 60 mm #1 (Thermo Fisher Scientific, Menzel-Gläser, catalog number: CS2460100 )
- Aluminium foil
- Plant pots 6 cm diameter (Volume 120 ml)
- Plants
Soybean (Glycine max Merr., cv. 29-I, Semillas Panorama, Colombia) plants of V2 (2nd trifoliate) stage and of comparable height were used - Bacterial strain
Burkholderia gladioli Lv-StA bacterial culture stored in 30% glycerol at -80 °C, previously isolated from accessory glands of a female L. villosa beetle (Flórez et al., 2017) - Molecular biology working kits
- MasterPureTM Complete DNA and RNA Purification Kit (Epicentre, catalog number: MC85200 )
- QuantiTect® Reverse Transcription Kit (QIAGEN, catalog number: 205311 )
- SYBR® Green Rotor-Gene PCR Kit (QIAGEN, catalog number: 204074 )
- InnuPREP Gel Extraction Kit (Analytik Jena, catalog number: 845-KS-5030050 )
- MasterPureTM Complete DNA and RNA Purification Kit (Epicentre, catalog number: MC85200 )
- Other reagents
- Sterile tap water (autoclaved)
- H2O treated with DEPC (Diethylpyrocarbonate) (Carl Roth, catalog number: K028.1 )
- Liquid nitrogen
- RNaseZap® Decontamination Solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9782 )
- Ethanol 100% (Carl Roth, catalog number: 5054.5 )
- H2O2 6% in 100% ethanol (Carl Roth, catalog number: 9681.4 )
- DAPI (4',6-Diamidino-2-phenylindol Dihydrochlorid) (Carl Roth, catalog number: 6335.1 )
- Ethanol 70%
- Milli-Q® water
- VECTASHIELD® (Vector Laboratories, catalog number: H-1000 )
- Peptone from soybean (AppliChem, catalog number: A2206 )
- Potassium phosphate dibasic (K2HPO4) (Carl Roth, catalog number: P749.1 )
- Glycerol (Carl Roth, Rotipuran®, catalog number: 3783.1 )
- Agar-Agar Kobe I (Carl Roth, catalog number: 5210.2 )
- Magnesium sulfate hydrate (MgSO4·xH2O) (Carl Roth, catalog number: 0261.1 )
- Tris/HCl (Carl Roth, Pufferan®, catalog number: 4855.2 )
- EDTA solution pH 8.0 (EUROCLONE, catalog number: EMR034500 )
- Triton® X-100 (Carl Roth, catalog number: 3051.3 )
- Sodium chloride (NaCl) (Carl Roth, catalog number: 3957.1 )
- Potassium chloride (KCl) (Carl Roth, catalog number: 6781.3 )
- Sodium phosphate dibasic (Na2HPO4) (Carl Roth, catalog number: P030.1 )
- Potassium phosphate monobasic (KH2PO4) (Carl Roth, catalog number: 3904.2 )
- Chloroform (Carl Roth, catalog number: 7331.2 )
- Acetic acid 100% (Carl Roth, catalog number: 6755.1 )
- Sodium dodecyl sulfate (SDS) (Carl Roth, catalog number: 4360.1 )
- Sterile tap water (autoclaved)
- Media and buffers (see Recipes)
- King’s B liquid and solid medium
- Low TE-buffer
- PBS-Tx buffer
- Carnoy’s fixative
- Hybridization buffer
- Washing buffer
- King’s B liquid and solid medium
Equipment
- Sticky insect tape (CONRAD, GLUPAC, model: GB011 )
- Class II biological safety cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: Safe 2020 , 1.2)
- Glass inoculation spreader
- Scissors
- Forceps
- Drying oven (Memmert, model: INB 200 )
- Vortex (Scilogex, model: MX-S , catalog number: 821200049999)
- Micropipettes 1 ml and 10 µl (Eppendorf, model: Research® plus, catalog numbers: 3120000062 and 3120000020 )
- Chlorophyll meter (Konica Minolta, model: SPAD-502Plus )
- Spectrophotometer UV/VIS (Eppendorf, model: BioPhotometer 6131 , catalog number: 6131 000.012)
- Heater mixing block (Biozym, Bioer Technology, model: MB-102 )
- Micro-centrifuge (Eppendorf, model: 5418 , catalog number: 5418000017)
- Real-time PCR cycler (QIAGEN, model: Rotor-Gene Q )
- NanoDropTM 1000 spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NDTM-1000 )
- Fluorescent microscope (ZEISS, model: AxioImager Z1 )
- Colour camera (ZEISS, model: AxioCam MRm )
Software
- SPSS (version 17.0)
- RStudio (version 0.98.1103)
- AxioVision software (Lite Edition 4.8.1, ZEISS)
Procedure
- Breeding and preparation of soybean plants
- Plant five soybean seeds per pot, in a total of 36 pots (180 seeds) and grow for seven days under greenhouse conditions (25 °C, 16:8 h light:dark cycle).
- Transfer plants to individual pots and grow for 21 days under greenhouse conditions.
- Select 36 plants of similar height and at developmental stage V2.
- Make a circular wound (5 mm diameter) in the central area of single leaflets of the first and second trifoliate leaves from each plant (Figure 1), using a robotic device that mimics insect herbivory damage (‘MecWorm’) (Mithöfer et al., 2005).
Note: Alternatively, a cork borer, a paper hole punch or a sterile blade can be used for wounding.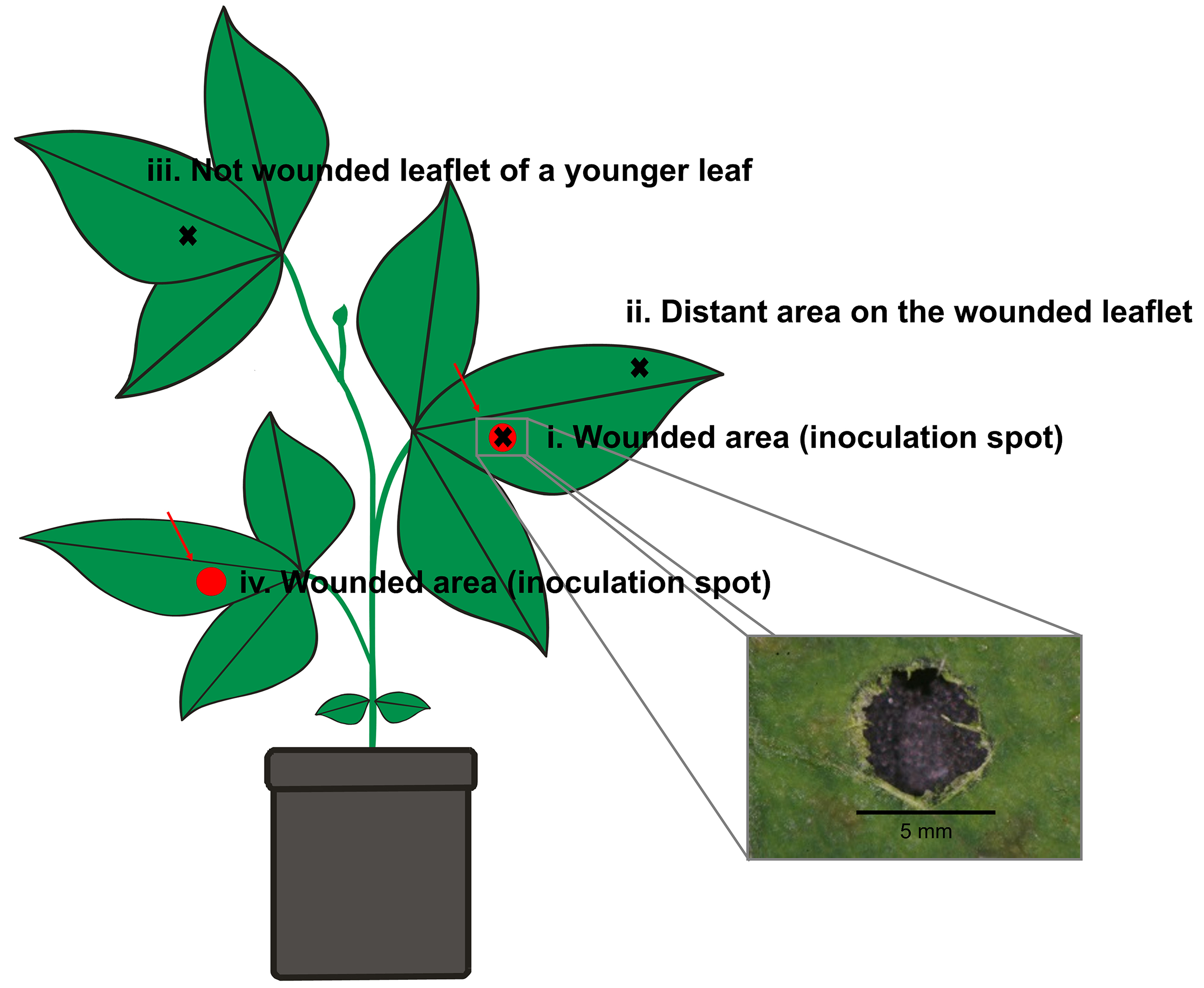
Figure 1. Schematic representation of the wounding and tissue sampling areas on a soybean plant. The scheme illustrates two wounded spots on the first and second trifoliate leaf (red regions), which had been inoculated at developmental stage V2 with B. gladioli Lv-StA in treated plants or sterile tap water in control plants. Black crosses indicate leaf areas later taken from developed soybean plants and used for FISH and quantitative PCR detection of B. gladioli Lv-StA. Note that wounding and infection were done at developmental stage V2 (only 2 fully developed trifoliate leaves present), while the scheme represents a plant at a later developmental stage, in which newer trifoliate leaves were also available for tissue sampling. - Use sticky insect traps between plants to prevent potential contact with insects and thereby cross contamination between plants.
- Arrange plants randomly and shuffle every second day under controlled light management (Figure 2).

Figure 2. Soybean plants organized in random order according to treatment and under controlled light conditions. The picture was acquired three weeks after inoculation. - Water plants every second day with tap water.
- Plant five soybean seeds per pot, in a total of 36 pots (180 seeds) and grow for seven days under greenhouse conditions (25 °C, 16:8 h light:dark cycle).
- Calibration curve for determining bacterial cell concentration using optical density
Note: Constructing a calibration curve is only necessary once for a specific spectrophotometer and bacterial strain, and should be done at least four days before initial plant inoculation considering the time required for CFU counting and preparation of the bacterial inoculum for infection.- Culture symbiotic B. gladioli Lv-StA in a 50 ml centrifuge tube containing 20 ml King’s B liquid medium (see Recipes) at 30 °C and constant shaking (200 rpm) for 24 h.
- Centrifuge the bacterial culture at 2,400 x g for 5 min. Discard the supernatant and resuspend the bacterial pellet in sterile tap water.
- Prepare three dilution series from 10-1 to 10-7 in 1.5 ml reaction tubes using sterile tap water (i.e., three times seven dilutions).
- Measure optical density (OD600) of each sample three times, and plate 50 µl per sample on Petri dishes containing King’s B medium agar.
- Incubate the Petri dishes at 30 °C for 48-72 h.
- Count colony forming units (CFU) of 3-day old cultures on King’s B medium plates (only CFU counts between 20 and 200 colonies per plate were considered).
- Generate a calibration curve using a linear regression of OD600 in relation to CFU count.
- Calculate the OD600 corresponding to 105 CFUs µl-1.
- Culture symbiotic B. gladioli Lv-StA in a 50 ml centrifuge tube containing 20 ml King’s B liquid medium (see Recipes) at 30 °C and constant shaking (200 rpm) for 24 h.
- Bacterial inoculum preparation and plant infection
Note: Bacterial inoculation was performed within two hours after wounding.- Culture symbiotic B. gladioli Lv-StA in King’s B liquid medium at 30 °C and constant shaking (200 rpm) for 24 h (i.e., one day before plant wounding and infection).
- Centrifuge the bacterial culture at 2,400 x g for 5 min. Discard the supernatant and resuspend the bacterial pellet in sterile tap water.
- Measure the OD600 of the bacterial suspension and dilute to 105 CFUs μl-1 using sterilized tap water.
- Inoculate 18 plants with 10 µl of bacterial suspension each by placing a drop with a micropipette on the two wounded areas of the trifoliate leaves for each plant (Figure 1).
- Apply 10 µl of sterilized tap water on the two corresponding wounds of each of the 18 control plants (Figure 1).
- Culture symbiotic B. gladioli Lv-StA in King’s B liquid medium at 30 °C and constant shaking (200 rpm) for 24 h (i.e., one day before plant wounding and infection).
- Monitoring of plant fitness
- Monitor plant height, as well as time to flowering, every second day until day 38 after infection (a total of 20 measurement time points including the day of inoculation).
- Observe potential symptoms of leaf necrosis and chlorosis (Figure 3).
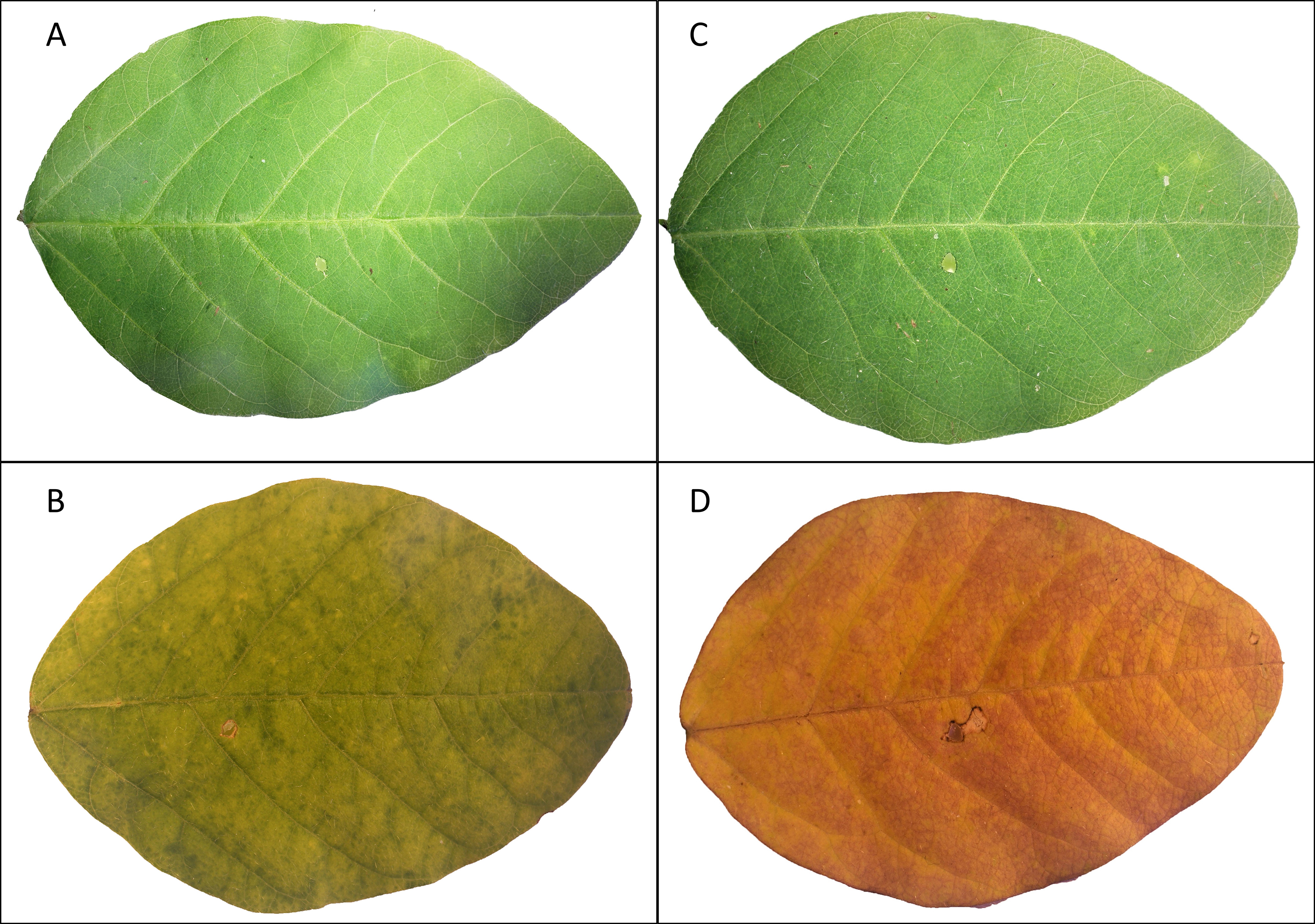
Figure 3. Chlorosis of single soybean leaflets treated with Burkholderia gladioli Lv-StA in comparison to a water control. Leaflets wounded and treated with sterile water are shown close after treatment (A) and 38 days after treatment (B), as well as leaflets infected with bacterial suspension close after treatment (C) and 38 days after being treated (D). - Measure the chlorophyll contents on three different spots of each wounded trifoliate leaflet every second day (triplicate measurements, not on the infection area) by using a chlorophyll meter SPAD-502Plus.
- Count the total number of seeds from all pods 38 days after infection.
- Also on day 38, dry the complete plant (above- and belowground parts) at 70 °C for 24 h and measure dry weight.
Important: See Note at the beginning of Procedure E
- Monitor plant height, as well as time to flowering, every second day until day 38 after infection (a total of 20 measurement time points including the day of inoculation).
- Quantification of infecting bacteria
Note: Harvest two leaflets from each plant at the end of the experiment (at day 38 after inoculation) before measuring the dry weight of the complete plant (see Step D5).- Cut pieces of leaflets (approximate area 1 x 1.5 cm) from different areas of each plant (see Figure 1) with sterile forceps and scissors and divide each sample into halves.
- Store half of each piece in sterile 1.5 ml tubes at -80 °C until further processing. Keep the other half of each piece for FISH analysis.
- Prepare sterile blunt 1 ml pipette tips as pestles. Flame the top of each tip shortly and then round it in 1.5 ml tubes containing DEPC treated H2O (Video 1).
Video 1. Preparation of pestles from 1 ml sterile tips - Weigh the stored leaflet pieces individually and then homogenize each sample in 1.5 ml tubes with the previously prepared pestles by pouring liquid nitrogen directly into each tube and crushing the tissue into powder (Video 2). If necessary for complete homogenization, add additional liquid nitrogen into the 1.5 ml tube and rapidly insert the pestle in the tube for grinding. While the pestle is not in use, you may partially close the tube (not tightly) to prevent powdered leaf tissue from spurting out of the tube.
Video 2. Plant tissue homogenization using liquid nitrogen - Extract nucleic acids using the MasterPureTM Complete DNA and RNA Purification Kit according to the manufacturer’s instructions. Perform centrifugation steps at 4 °C and resuspend DNA and RNA in Low TE-buffer (see Recipes).
- Perform reverse transcription on ice using the QuantiTect® Reverse Transcription Kit including a genomic DNA removal step and a corresponding control, following the manufacturer’s instructions.
- Conduct quantitative RT-PCR on the corresponding cDNA on a 167 bp fragment using primers Burk16S_StAG_F and Burk 3.1_R (Table 1) in a RotorGene®-Q cycler device following the protocol described for the Rotor-Gene SYBR Green PCR Kit.
Table 1. Primers and probes used for amplification and fluorescence in situ hybridization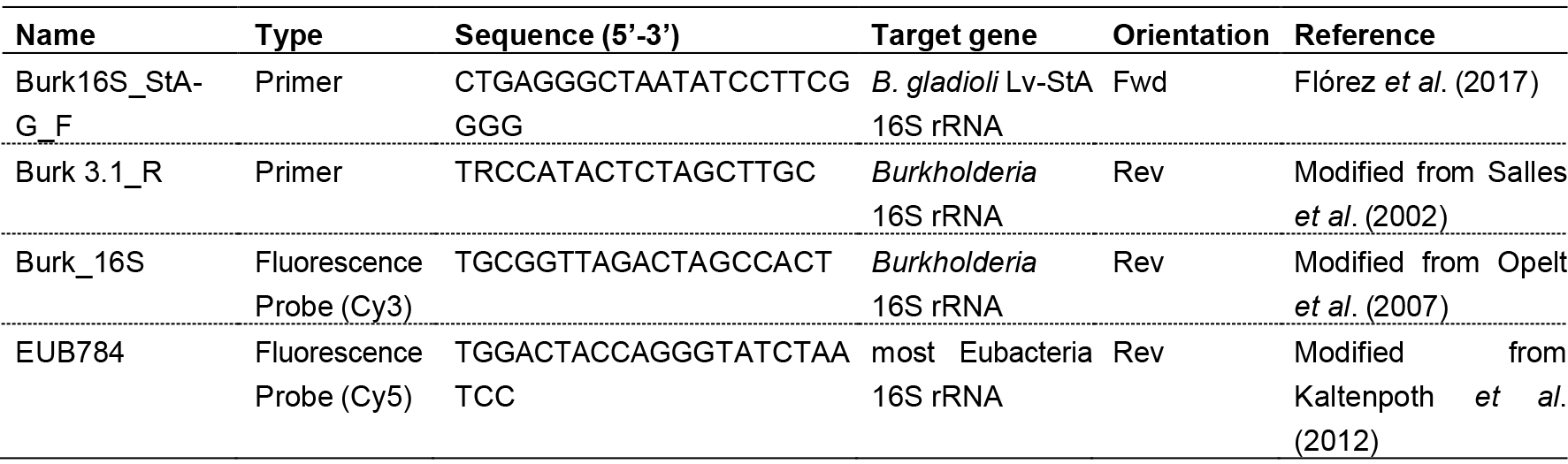
- Carry out a quantitative RT-PCR consisting of an initial denaturation step at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 10 sec, primer annealing at 65 °C for 30 sec and elongation at 72 °C for 20 sec. Include a melting curve with a temperature gradient from 60 °C to 99 °C within 4.25 min.
- DNA standards for quantification of the target fragments were obtained previously by PCR amplification and product recovery using the innuPREP Gel Extraction Kit. Purified DNA concentration was measured using a NanoDropTM 1000 spectrophotometer. Standards were used as a tenfold dilution series ranging from 10-1 to 10-8 ng µl-1.
- All solutions used for RNA extraction were prepared in DEPC (Diethylpyrocarbonate) treated H2O and RNaseZap® Decontamination Solution was used in order to avoid RNase contamination of equipment and workspaces.
- Cut pieces of leaflets (approximate area 1 x 1.5 cm) from different areas of each plant (see Figure 1) with sterile forceps and scissors and divide each sample into halves.
- Visualization of the infecting bacteria by FISH
- Cut and thinly slice the stem tissue of different plant regions (between 1st and 2nd trifoliates and above the 2nd trifoliate) with a razor blade in approximately 0.5 mm longitudinal sections.
- Fixate the sliced stem tissue and the half piece of leaflet recovered previously (see Step E1) at room temperature (RT) for 24 h in Carnoy’s fixative (see Recipes) and store at RT until further use.
- Wash samples at RT in 100% ethanol for 2 h and then bleach in 6% H2O2 (diluted in ethanol) for approximately 14 days. Change the bleach solution every third day.
- Wash the samples at RT in 100% ethanol (3 x 2 h), in 70% ethanol (2 x 1 h) and in PBS-Tx buffer (4 x 0.5 h, see Recipes).
- Hybridize the samples at 55 °C for 24 h in 1.5 ml tubes containing 270 µl hybridization buffer (see Recipes) with 15 µl (50 ng µl-1) of the Cy5-labeled general eubacterial probe EUB784, 15 µl (50 ng µl-1) of the Cy3-labeled Burkholderia specific probe Burk_16S (Table 1) and additionally 3 µl DAPI (0.5 mg ml-1) for host cell counterstaining.
- Wash samples at 55 °C in washing buffer (2 x 2 h, see Recipes) and then incubate samples in distilled H2O and a small amount (1-2 drops) of VECTASHIELD® for 2 h at room temperature.
- Transfer samples to a glass slide, cover with VECTASHIELD® and a cover slip, and observe samples using a fluorescence microscope (Figure 4).
- Step F3 is light sensitive and should be performed under dark or dim light conditions. We recommend covering the samples with aluminium foil or similar during incubation. Decolorization with bleaching solution is essential to avoid auto-fluorescence of the plant tissue.
- Steps F5-F7 involve the manipulation of fluorescent probes, therefore we recommend working under dark or dim light to minimize bleaching.
- During and until the final washing step with distilled H2O (Steps F5 and F6), it is essential that samples do not dry out completely, as the subsequent removal of unhybridized probe might be compromised.
- Some FISH probes designed for eubacteria might also hybridize to the plant chloroplasts (e.g., EUB388: 5’-GCTGCCTCCCGTAGGAGT-3’). The probe EUB784 was convenient in this sense, as no hybridization to chloroplasts was observed in silico nor in situ.
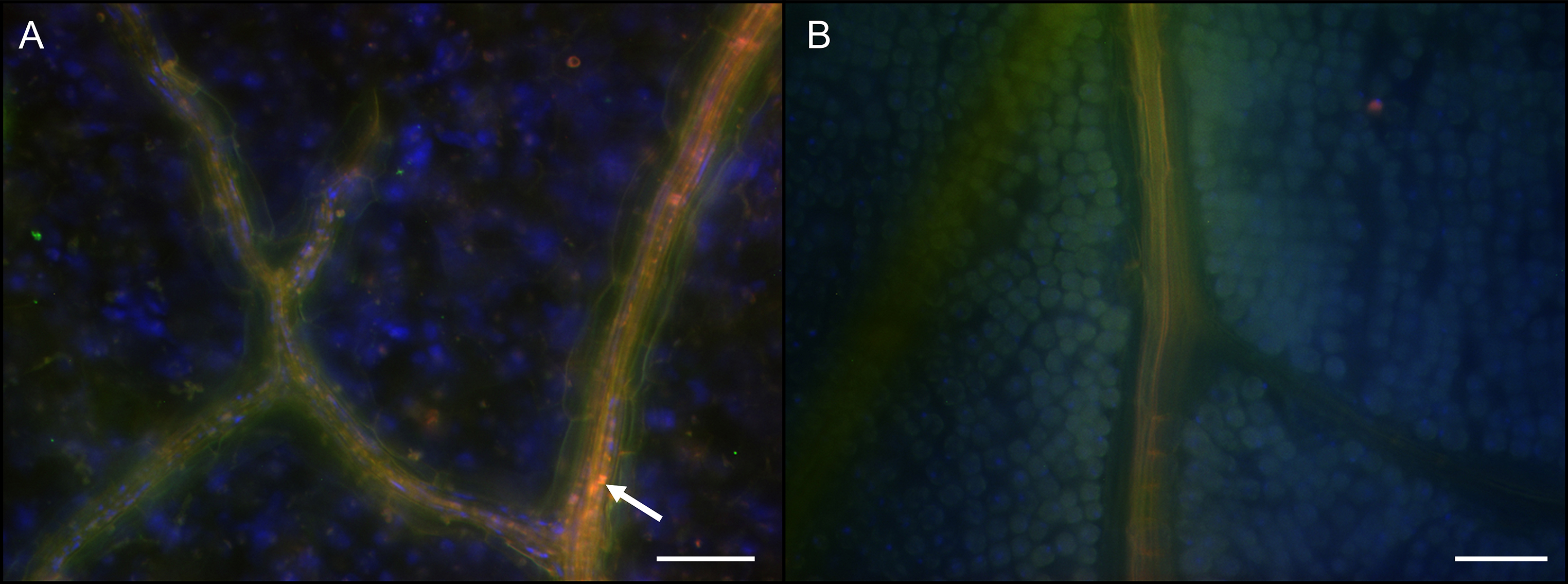
Figure 4. Localization of Burkholderia gladioli cells in the soybean plant vascular system. Whole mount FISH on soybean leaf tissue showing an overlay image of hybridization with Burk_16S-Cy3 (red), EUB784-Cy5 (green) and host cell nuclei staining with DAPI (blue). The white arrow indicates Burkholderia cells in the vascular system of an infected soybean plant leaflet (A). No Burkholderia cells were observed in control plants (B). The scale bars represent 50 µm. - Cut and thinly slice the stem tissue of different plant regions (between 1st and 2nd trifoliates and above the 2nd trifoliate) with a razor blade in approximately 0.5 mm longitudinal sections.
Data analysis
Statistical analyses were performed using SPSS (version 17.0). Seed output (number of seeds) and first flowering events were compared between B. gladioli infected plants and the uninfected controls using the Mann-Whitney U-test, given the non-normal distribution of the data. RStudio (version 0.98.1103) was used for linear mixed model (LMM) and generalized additive model (GAM) analysis using time as random factor and chlorophyll and treatment or height and treatment as fixed factors. The quantitative PCR data on Burkholderia abundance in different plant locations upon infection were analyzed using an ANOVA and Tukey’s post hoc tests after confirming the normal distribution of the data and homogeneity of variances. Results on seed output and Burkholderia quantification in plant tissues have been published previously (Flórez et al., 2017).
Recipes
- King’s B medium
20 g peptone
1.5 g K2HPO4
10 ml glycerol
15 g agar (only for solid media)
1 L double distilled water
Autoclave, then add 5 ml of sterile 1 M MgSO4 - Low TE-buffer
10 mM Tris/HCl (in Milli-Q® water)
0.1 mM EDTA (in Milli-Q® water) - PBS-Tx buffer
3 g Triton® X-100
8 g NaCl
0.2 g KCl
1.4 g Na2HPO4
0.3 g KH2PO4
1 L double distilled H2O - Carnoy’s fixative
6 parts ethanol 100%
3 parts chloroform 100%
1 part acetic acid 100% - Hybridization buffer
900 mM NaCl
0.01% sodium dodecyl sulfate (SDS)
20 mM Tris/HCl pH 8.0 - Washing buffer
5 mM EDTA
100 mM NaCl
0.1% SDS
20 mM Tris/HCl pH 8.0
Acknowledgments
This protocol was adapted from (Flórez et al., 2017) and was carried out at the Max Planck Institute for Chemical Ecology (Jena, Germany). A specialized device for mechanical plant wounding previously developed by Dr. Axel Mithöfer et al. (2005) was used. We thank the Greenhouse team at the Max Planck Institute for Chemical Ecology for soybean cultivation and the Max-Planck-Society for funding. The authors have no conflict of interest or competing interests to declare.
References
- Flórez, L. V. and Kaltenpoth, M. (2017). Symbiont dynamics and strain diversity in the defensive mutualism between Lagria beetles and Burkholderia. Environ Microbiol 19(9): 3674-3688.
- Flórez, L. V., Scherlach, K., Gaube, P., Ross, C., Sitte, E., Hermes, C., Rodrigues, A., Hertweck, C. and Kaltenpoth, M. (2017). Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat Commun 8: 15172.
- Frago, E., Dicke, M. and Godfray, H. C. (2012). Insect symbionts as hidden players in insect-plant interactions. Trends Ecol Evol 27(12): 705-711.
- Frank, S. A. (1997). Models of symbiosis. Am Nat 150 Suppl 1: S80-99.
- Gilbert, S. F., Sapp, J. and Tauber, A. I. (2012). A symbiotic view of life: we have never been individuals. Q Rev Biol 87(4): 325-341.
- Kaltenpoth, M., Yildirim, E., Gurbuz, M. F., Herzner, G. and Strohm, E. (2012). Refining the roots of the beewolf-Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl Environ Microbiol 78(3): 822-827.
- Mithöfer, A., Wanner, G. and Boland, W. (2005). Effects of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiol 137(3): 1160-1168.
- Opelt, K., Berg, C., Schonmann, S., Eberl, L. and Berg, G. (2007). High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J 1(6): 502-516.
- Salles, J. F., De Souza, F. A. and van Elsas, J. D. (2002). Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl Environ Microbiol 68(4): 1595-1603.
- Stammer, H. J. (1929). Die Symbiose der Lagriiden (Coleoptera). Zoomorphology 15(1/2): 1-34.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gaube, P., Kaltenpoth, M. and Flórez, L. V. (2017). Infection of Soybean Plants with the Insect Bacterial Symbiont Burkholderia gladioli and Evaluation of Plant Fitness. Bio-protocol 7(24): e2663. DOI: 10.21769/BioProtoc.2663.
Category
Plant Science > Plant immunity > Host-microbe interactions
Microbiology > Microbe-host interactions > Bacterium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


.jpg)









