- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A General Method for Intracellular Protein Delivery through ‘E-tag’ Protein Engineering and Arginine Functionalized Gold Nanoparticles
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2661 Views: 8284
Reviewed by: Renate WeizbauerPia GiovannelliYang Fu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Induction and Detection of Acrosomal Exocytosis in Human Spermatozoa
Shenae L. Cafe [...] Brett Nixon
Jul 20, 2020 4778 Views

A New Method for Studying RNA-binding Proteins on Specific RNAs
Weiping Sun [...] Min Zhuang
May 20, 2021 10583 Views

Flow Cytometry Analysis of Microglial Phenotypes in the Murine Brain During Aging and Disease
Jillian E. J. Cox [...] Sarah R. Ocañas
Jun 20, 2024 3569 Views
Abstract
In this protocol, we describe a method for direct cytosolic protein delivery that avoids endosomal entrapment of the delivered proteins. We achieved this by tagging the desired protein with an oligo glutamic acid tag (E-tag), and subsequently using carrier gold nanoparticles to deliver these E-tagged proteins. When E-tagged proteins and nanoparticles were mixed, they formed nanoassemblies, which got fused to cell membrane upon incubation and directly released the E-tagged protein into cell cytosol. We used this method to deliver a wide variety of proteins with different sizes, charges, and functions in various cell lines (Mout et al., 2017).
To use this protocol, the first step is to generate the required materials (gold nanoparticles, recombinant E-tagged proteins). Laboratory-synthesis of gold nanoparticles has been previously described (Yang et al., 2011). Desired E-tagged proteins can be cloned from the corresponding genes, and expressed and purified using standard laboratory procedures. We will use E-tagged green fluorescent protein (GFP) as a reference protein here. Users can simply insert an E-tag into their protein of interest, at either terminus. To achieve maximum delivery efficiency, we suggest users testing different length of E-tags. For example, we inserted E = 0 to 20 (E0 means no E-tag insertion, and E20 means 20 glutamic acids insertion in a row) to most of the proteins we tested, and screened for optimal E-tagged length for highest delivery efficiency. E10-tagged proteins gave us the highest delivery efficiency for most of the proteins (except for Cas9, where E20 tag showed highest delivery efficiency).
Once these materials are ready, it takes about ~10 min to make the E-tagged protein and nanoparticle nanoassemblies, which are immediately used for delivery. Complete delivery (~100% for GFP-E10) is achieved in less than 3 h.
Background
Intracellular delivery of exogenous proteins into cells is crucial for cellular imaging and diagnosis, therapeutic development, genome engineering and synthetic biology applications (Fu et al., 2014). Many of the applications in cellular engineering and imaging (such as genome editing, cellular imaging) require delivering exogenous proteins, as mammalian cells do not have genes for those proteins. However, access to the whole cytoplasm by the delivered protein remains elusive. A major hurdle in cytoplasmic protein delivery is the endosomal entrapment of the delivered cargo: nanocarrier-based delivery methods result in only a fraction of the entrapped cargo (often ~1%) escaping into the cytosol (Stewart et al., 2016). Additionally, protease-mediated degradation and exocytosis of the remaining entrapped cargo proteins make these strategies ultimately inefficient. Delivery through membrane disruption methods can provide efficient cytosolic protein delivery; however, these methods generally require additional osmolytic surfactants (Erazo-Oliveras et al., 2014), hypertonic agents (D’Astolfo et al., 2015), or mechanical distortion techniques (Han et al., 2015) that may be harmful for the cells. Our protocol provides an approach for direct cytosolic delivery of a desired protein for applications including cellular imaging and basic cell biology research (Mout et al., 2017).
Materials and Reagents
- Round bottom 35 mm confocal dish (MATTEK, catalog number: P35G-0-14-C )
- 24-well plates (Corning, Costar®, catalog number: 3524 )
- Sterile tubes (0.6, 1.5, and/or 2.0 ml) (Fisherbrand)
- Sterile pipette tips (white, yellow and blue tips)
- Cell lines (i.e., HeLa)
- 1x Phosphate buffered saline (PBS) (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- Plain DMEM media (No serum and antibiotics) (Thermo Fisher Scientific, GibcoTM, catalog number: 10567014 )
- Appropriate media for cell culture (i.e., HeLa cells, DMEM, Thermo Fisher Scientific, GibcoTM, catalog number: 10567014 )
- Alexa Fluor 488 NHS Ester (Thermo Fisher Scientific, InvitrogenTM, catalog number: A20000 )
- Stock solution of ArgNP gold nanoparticles (~50 μM in water, laboratory synthesized)
- E-tagged proteins (laboratory purified)
Equipment
- Pipettes (P2, P10, P20, P200, P1000)
- BSL-2 safety cabinet for cell culture
- Cell culture incubator at 5% CO2 and 37 °C
- Confocal microscope (any brand)
- Flow cytometer (i.e., BD, model: LSR II )
Procedure
There are two key steps for successful high efficient cytosolic protein delivery. First, the length of E-tag determines the delivery efficiency. For each protein, the length may be different. Therefore, users are suggested to make their protein of interest with at least few different E-tags (of different E-tag length, for example, E5, E10, E15, and E20). For most average molecular weight proteins (MW < 50 kDa), E10 tag gives high efficient delivery, however, for large proteins such as Cas9 (MW = 160 kDa) E20 tag gives best delivery. Second, the molar ratio of ArgNPs to E-tagged protein determines the delivery efficiency. Again, for each protein, this ratio may be different. Therefore, users are also suggested to test few different molar ratios of ArgNPs/E-tagged protein of their interest to find out the best delivery. From our experience, for most E-tagged proteins, one of the following molar ratios gave highest delivery efficiency: [ArgNP]/[E-tagged protein] = 1:0.5; 1:1; 1:2; 1:3. However, users are suggested to test any other ratios as they may think appropriate.
In the following protocol (Figure 1), we use GFP-E10 to demonstrate the assembly formation and delivery process. However, as mentioned above, users are requested to identify the optimum E-tag length and ‘working molar ratio’ for their protein of interest using the same procedure as described for GFP-E10. Additionally, E-tagged proteins should be fluorophore-labeled to assess delivery efficiency by confocal microscopy imaging, if the protein is a non-fluorescent protein. [For Alexa Fluor 488 NHS Ester, a labeling protocol can be found on manufacturer’s website.]
Final working nanoassembly concentration is 250 nM of ArgNPs and 750 nM of GFP-E10, which is at 1:3 molar ratio of ArgNPs/GFP-E10. The total volume of the nanoassembly samples required for delivery depends on the kind of cell culture plate used. We generally use 1 ml for round bottom 35 mm confocal dish, 500 μl for 24-well plates, per well. Therefore, the nanoassemblies should be prepared and scaled up according to users need. The following calculation is for one sample in 24-well plate (i.e., 500 μl total volume). Additionally, the following protocol is for HeLa cells only. Nevertheless, we have used a wide variety of cell lines to demonstrate our delivery platform. Some cell lines that we successfully tested are: human embryonic kidney cells (HEK), mammary epithelial cells (MCF-7), mouse macrophage (RAW 264.7), human ovarian cancer cells (SKOV-3), and T-lymphocyte cells (Jurkat).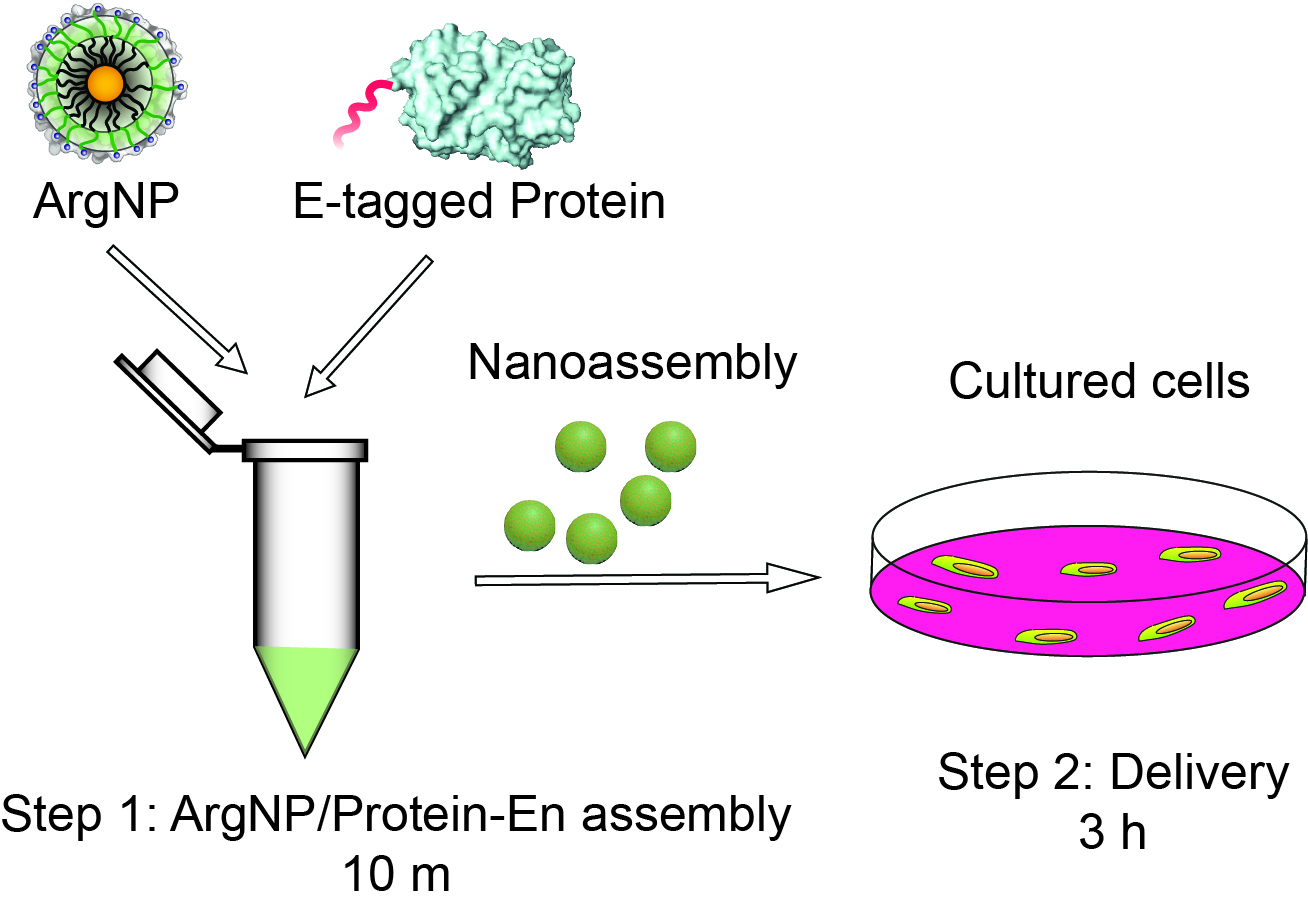
Figure 1. Schematic overview of the protocol. Step 1: Formation of ArgNPs/E-tagged protein nanoassembly (takes 10 min), and Step 2: E-tagged protein delivery (takes up to 3 h).
Day 1
Seed cells on 24-well plate at a cell density of 0.8-1 x 105 cells/well (For confocal dish, seed 2.4 x 105 cells per dish). Let the cells grow for 24 h at 5% CO2 and 37 °C in a cell culture incubator.
Day 2
- Step 1: ArgNPs/E-tagged protein nanoassembly preparation
- This step (Step 1) should be performed 10 min before the delivery experiment (Step 2).
- Add 100 μl of 1x PBS into a sterile 1.5 ml tube.
- Add 2.5 μl of ArgNPs (50 μM stock) into it.
Note: The final working concentration of ArgNPs is 250 nM, and the total working sample volume is 500 μl. If the stock concentration of ArgNPs is different, please adjust the amount (volume) of nanoparticle to be added. Also, scale up or down based on the total working volume of the assembly sample. - Then add 7.5 μl of GFP-E10 (50 μM stock) into the ArgNPs solution and mix well. Here, the final working concentration of GFP-E10 is 750 nM at a [ArgNPs]/[GFP-E10] molar ratio = 1:3 (250:750 nM). Again, adjust the volume of GFP-E10 or E-tagged protein of interest based on stock solution concentration and required molar ratio.
Note: At this step, users can choose their protein of interest with different E-tag length and a few [ArgNPs]/[E-tagged protein] molar ratios to find out maximum cytosolic delivery. - Incubate the complex at room temperature for 10 min.
- The nanoassembly is ready for the next step, i.e., delivery.
- This step (Step 1) should be performed 10 min before the delivery experiment (Step 2).
- Step 2: Delivery
- While the nanoassembly was incubated for 10 min, users should prepare the cells for delivery in the meantime.
Note: Don’t incubate for more than 10 min, as this may cause aggregate formation instead of well-defined assemblies. - Wash the cells with 1x PBS, twice.
- Add ~400 μl of DMEM plain media (or any media of interest) into the nanoassembly solution in the tube. Mix well by gently pipetting up and down.
Note: Media with serum should be strictly avoided at this step; media with serum may require further optimization such as the length of E-tag, the ratio of nanoparticles to E-tagged protein, and incubation time. - Add the whole (500 μl) sample into the washed-cells.
- Incubate the cells in appropriate condition (usually 5% CO2 and 37 °C) for 3 h. Complete protein delivery should be achieved after 3 h of incubation.
- Wash away the media after 3 h, and replace with fresh desired media (DMEM with 10% serum and 1% antibiotics, and any supplements if needed).
- While the nanoassembly was incubated for 10 min, users should prepare the cells for delivery in the meantime.
Data analysis
- Two types of data assessments are performed to evaluate efficient cytosolic protein delivery. First, thorough distribution of the protein in the cytoplasm (using confocal microscopy imaging) would indicate effective delivery, whereas, punctate distribution would mean endosomal entrapment of proteins (Figures 2A and 2B). Second, the percentage of transduced cells can be assessed by using either confocal microscopy imaging or flow cytometry analysis (Figure 2C) (For detailed procedure for confocal microscopy and flow cytometry, please see Mout et al., 2017). Note that, the protein of interest must be fluorophore-labeled, or a fused fluorescent protein be used for the above imaging and quality assessment purposes.
- Following are a few representative results showing effective cytosolic delivery of GFP-E10. To compare the effectiveness of our strategy with other methods that suffer from endosomal entrapment of the delivered protein, we also provided an image showing (+36)-GFP delivery that predominately gets trapped in the endosomes (Cronican et al., 2010). Likewise, percentage GFP delivery was compared among GFP-E0, GFP-E5, and GFP-E10 using flow cytometry analysis. The result shows highest delivery efficiency for GFP-E10. Please see Mout et al., 2017 for more details.
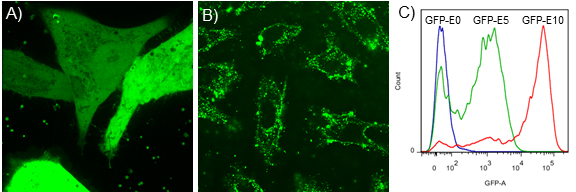
Figure 2. Representative data showing efficient protein delivery vs. ineffective delivery. Confocal microscopy images showing examples of effective cytosolic delivery of GFP-E10 (A), versus endosomal delivery of (+36)-GFP in HeLa cells (B). Note that, GFP-E10 was delivered using above protocol at a molar ratio of [ArgNPs]/[GFP-E10] = 1:3 (250:750 nM). For comparison purpose, we delivered (+36) GFP following a reported method (Cronican et al., 2010). C. Flow cytometric determination of delivery efficiency of E-tagged GFP with different tag length. As shown in the figure, GFP-E10 exhibited highest deliver efficiency.
Notes
Every batch of ArgNPs can be slightly deferent in terms of surface ligand coverage and therefore users are suggested to test different molar ratio of ArgNPs/E-tagged protein to find out optimum ratio for maximum delivery.
Acknowledgments
This research was supported by the NIH (GM077173), and NSF (CHE-1307021). This protocol was adapted from Mout et al., 2017.
The authors declare the following competing financial interest(s): V.M.R and R.M. submitted a non-provisional patent to USPTO (Application number PCT/US2016/015711) on the invention.
References
- Cronican, J. J., Thompson, D. B., Beier, K. T., McNaughton, B. R., Cepko, C. L. and Liu, D. R. (2010). Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. ACS Chem Biol 5(8): 747-752.
- D’Astolfo, D. S., Pagliero, R. J., Pras, A., Karthaus, W. R., Clevers, H., Prasad, V., Lebbink, R. J., Rehmann, H. and Geijsen, N. (2015). Efficient intracellular delivery of native proteins. Cell 161(3): 674-690.
- Erazo-Oliveras, A., Najjar, K., Dayani, L., Wang, T. Y., Johnson, G. A. and Pellois, J. P. (2014). Protein delivery into live cells by incubation with an endosomolytic agent. Nat Methods 11(8): 861-867.
- Fu, A., Tang, R., Hardie, J., Farkas, M. E. and Rotello, V. M. (2014). Promises and pitfalls of intracellular delivery of proteins. Bioconjug Chem 25(9): 1602-1608.
- Han, X., Liu, Z., Jo, M. C., Zhang, K., Li, Y., Zeng, Z., Li, N., Zu, Y. and Qin, L. (2015). CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv 1(7): e1500454.
- Mout, R., Ray, M., Tay, T., Sasaki, K., Yesilbag Tonga, G. and Rotello, V. M. (2017). General strategy for direct cytosolic protein delivery via protein-nanoparticle co-engineering. ACS Nano 11(6): 6416-6421.
- Stewart, M. P., Sharei, A., Ding, X., Sahay, G., Langer, R. and Jensen, K. F. (2016). In vitro and ex vivo strategies for intracellular delivery. Nature 538(7624): 183-192.
- Yang, X. C., Samanta, B., Agasti, S. S., Jeong, Y., Zhu, Z. J., Rana, S., Miranda, O. R. and Rotello, V. M. (2011). Drug delivery using nanoparticle-stabilized nanocapsules. Angew Chem Int Ed Engl 50(2): 477-481.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mout, R. and Rotello, V. M. (2017). A General Method for Intracellular Protein Delivery through ‘E-tag’ Protein Engineering and Arginine Functionalized Gold Nanoparticles. Bio-protocol 7(24): e2661. DOI: 10.21769/BioProtoc.2661.
Category
Biochemistry > Protein > Labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










