- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring the Endocytic Recycling of Amyloid Precursor Protein (APP) in Neuro2a Cells
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2635 Views: 8138
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Immunostaining Protocol: P-Smad2 (Xenograft and Mice)
Alexandre Calon [...] Eduard Batlle
May 5, 2014 9024 Views

Combined in situ Hybridization/Immunohistochemistry (ISH/IH) on Free-floating Vibratome Tissue Sections
Manuel E. Lopez
Sep 20, 2014 15474 Views

Non-separate Mouse Sclerochoroid/RPE/Retina Staining and Whole Mount for the Integral Observation of Subretinal Layer
Sung-Jin Lee and Soo-Young Kim
Jan 5, 2021 4451 Views
Abstract
The established primary trigger of Alzheimer’s disease’s is β-amyloid (Aβ) (Mucke and Selkoe, 2012). Amyloid precursor protein (APP) endocytosis is required for Aβ generation at early endosomes (Rajendran and Annaert, 2012). APP retention at endosomes also depends on its recycling back to the plasma membrane (Koo et al., 1996; Ubelmann et al., 2017). The following recycling assay has been optimized to assess APP recycling by live murine Neuro2a cells, a neuroblastoma cell line (Ubelmann et al., 2017).
Keywords: APPBackground
Aβ42 accumulation is a primary trigger of Alzheimer’s disease. APP endocytosis is required for Aβ42 generation (Koo and Squazzo, 1994; Grbovic et al., 2003; Cirrito et al., 2008; Rajendran et al., 2008). Upon endocytosis, APP can recycle back to the plasma membrane likely escaping processing at endosomes. While many studies have characterized APP endocytosis, mechanisms that regulate APP recycling need to be established. A robust, sensitive and quantitative assay is thus necessary. APP recycling has been assessed qualitatively by immunofluorescence and quantitatively using bulk biotinylation of surface proteins followed by a chase for endocytosis and a chase for recycling after stripping or blocking of surface proteins (Koo et al., 1996; Yamazaki et al., 1996; Chaufty et al., 2012).
We developed a methodology for following and measuring APP recycling in Neuro2a cells using classical immunofluorescence and semi-quantitative cell biology methods. Our assay relies on the transient expression of APP tagged with the red fluorescent protein (RFP), to detect and quantify the cellular pool of APP and on using an antibody against the ectodomain of APP, to selectively detect and quantify the trafficking of the surface pool of APP.
Materials and Reagents
- Circular glass coverslips, 13 mm (VWR, Marienfeld, catalog number: 630-1597 )
Note: Autoclaved, pre-washed with 40% ethanol/60% HCl 1 h at RT and washed 4 times, 15 min each, with Milli-Q water at RT.
- Superfrost glass slides (MENZEL GERHARD, catalog number: 2586E )
- 24-well dishes (SARSTEDT, catalog number: 83.1836 ) for mammalian cell culture
- Plastic Pasteur pipette (SARSTEDT, catalog number: 86.1171 )
- Parafilm (Fisher Scientific, catalog number: 11782644)
Manufacturer: Bemis, Parafilm, catalog number: PM999 .
- Neuroblastoma Neuro2a cells (ATCC, catalog number: CCL-131 )
- APP-RFP plasmid (Szodorai et al., 2009) (S. Kins, University of Kaiserslautern)
- Culture medium: Dulbecco’s modified Eagle medium (DMEM)-GlutaMAX (Thermo Fisher Scientific, GibcoTM, catalog number: 10566016 ) with 10% fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F0804 ); 100 U/ml penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Lipofectamine 2000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019 )
- Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985062 )
- Lipofectamine RNAiMax (Thermo Fisher Scientific, InvitrogenTM, catalog number: 13778150 )
- Murine Anti-APP N-terminal monoclonal antibody (22C11) (Merck, catalog number: MAB348 )
- HEPES (1 M) (Thermo Fisher Scientific, GibcoTM, catalog number: 15630080 )
- Phosphate buffer saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010031 )
- Donkey anti-mouse Alexa 488 (Thermo Fisher Scientific, catalog number: A-21202 )
- Coverslip-slide Mounting solution (FluoroMount-G) (Southern Biotech, catalog number: 0100-01 )
- Saponin (Sigma-Aldrich, catalog number: 47036 )
- Sodium chloride (NaCl) (NZYTech, catalog number: MB15901 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- Acetic acid (Merck, catalog number: 1.00063.2511 )
- Acid-stripping buffer (see Recipes)
- Fixative solution (see Recipes)
Equipment
- CO2 incubator for primary cell culture (BINDER, model: CB 160 )
- Counting chamber (Belden, Hirschmann, catalog number: 8100103 )
- Epifluorescence upright microscope Z2 (Carl Zeiss, model: Axio Imager Z2 ) equipped a 60x NA-1.4 oil immersion objective and an AxioCam MRm CCD camera (Carl Zeiss)
Software
- ImageJ 2.0 v1.51n software (free download from http://rsb.info.nih.gov/ij/)
- GraphPad Prism 6 (https://www.graphpad.com/scientific-software/prism/)
Procedure
Note: The APP recycling assay is performed on Neuro2a cells grown on glass coverslips.
- Cell culture and transfection (Day 1, Day 2)
- Place one washed coverslip in the bottom of each well of a 24-well plate.
- Plate Neuro2a cells (30,000 cells per well of 24-well plate; adjust the cell density so that Neuro2a cells do not reach confluence at the day of the recycling assay) in culture medium (see Reagents) at 37 °C in a 5% CO2 and 20% O2 humidified incubator (Day 1). We recommend using two coverslips per condition.
Note: Most cells will grow adherent to the glass coverslip.
- For expression of APP-RFP cDNA, 60-70% confluent Neuro2a cells are transiently transfected with 0.5 µg cDNA: 0.5 µl Lipofectamine 2000 mix in 25 µl Opti-MEM per well of 24-well plate with 250 µl of antibiotics-free complete medium. (Day 2)
- Replace transfection medium 2 h post-transfection with complete culture medium at 37 °C to avoid the excessive expression of APP-RFP (Day 2).
- (Optional) siRNA transfection for knockdown analysis with Lipofectamine RNAiMax (Thermo Fisher Scientific) according to manufacturer’s protocol 72 h prior to the recycling assay (Day 2).
- Place one washed coverslip in the bottom of each well of a 24-well plate.
- APP recycling assay in Neuro2a cells expressing APP-RFP by the following steps (Figure 1) (Day 3)
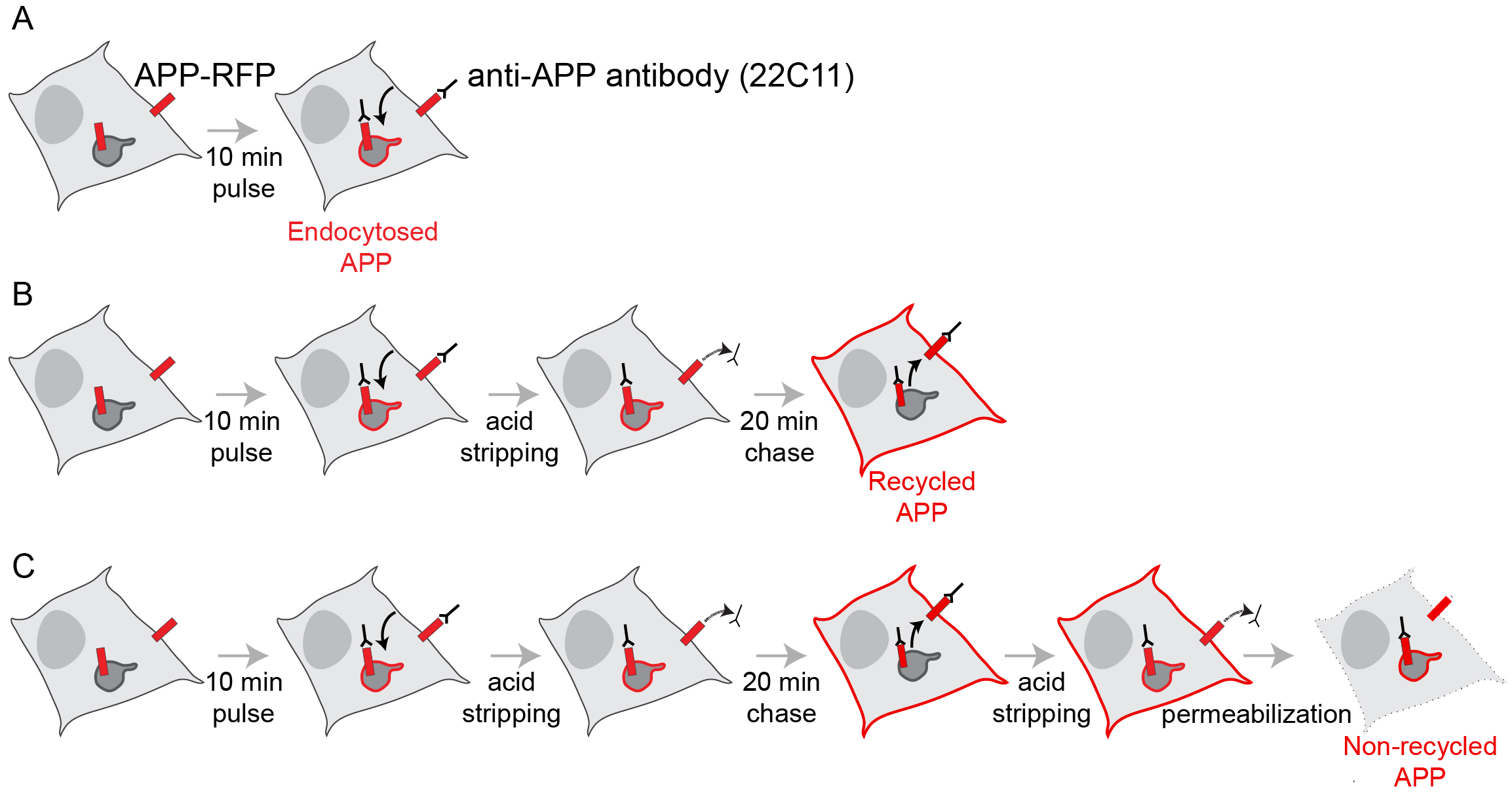
Figure 1. Schematic of protocols to monitor APP endocytosis, APP recycling and non-recycling APP. A. Pulse with anti-APP antibody for 10 min; B. Upon 10 min pulse, non-endocytosed APP at the surface is removed with acid-stripping buffer, and recycled APP at the surface is detected in non-permeabilized cells after a 20 min chase; C. Upon the 20 min chase, the non-recycled pool of APP is detected upon acid-stripping and permeabilization.
- Remove cell culture media with a plastic Pasteur pipette, add 500 µl serum-free culture medium at 37 °C and incubate cells for 30 min at 37 °C in cell culture incubator.
Note: All pipetting should be done slowly against the wall of the well without touching the cells.
- Dilute 0.25 µl of anti-APP antibody (22C11; 0.25 µg/µl stock concentration) into 25 µl culture medium with 10 mM HEPES (0.25 µl of stock solution at 1 M pH 7.2-7.5) per coverslip.
- Place one 25 µl droplet of diluted anti-APP antibody per glass coverslip onto Parafilm stretched on a 24 wells plate lid.
- Carefully place the coverslips over the droplets with cells facing the antibody solution and incubate them at 37 °C for 10 min in cell culture incubator. Cover the reaction to avoid evaporation. This step allows for anti-APP antibody binding to cell surface APP and subsequent endocytosis.
(Optional) For a surface labeling control coverslip, carefully transfer the coverslips over the droplets with cells facing the antibody solution and incubate them at 37 °C for 4 min in cell culture incubator. Wash each coverslip by dipping in PBS at 37 °C for 4 s. Place coverslips back in a 24-well plate and add 250 µl of 4% PFA fixative solution for 10 min at room temperature (RT). After washing 3 times with PBS (RT) proceed for detection of anti-APP. This optional step allows for anti-APP antibody binding to cell surface APP.
- Wash each coverslip by dipping in PBS at 37 °C for 4 s.
(Optional) For controlling the pool of endocytosed APP (see Figure 1A) proceed for fixation by placing the control coverslip(s) back in the 24-well plate and add 250 µl of 4% PFA fixative solution for 10 min at room temperature (RT). After washing 3 times with PBS (RT) proceed for detection of anti-APP.
- For assaying the pool of recycled APP bound to anti-APP (see Figure 1B) is necessary to:
- Remove of surface anti-APP antibody bound to non-endocytosed APP by dipping the corresponding coverslips into an acid-stripping buffer (see Recipes) for 5 s and careful washing by dipping the coverslips in PBS at 37 °C for 4 s.
- Place the coverslips back in a 24-well plate containing pre-warmed culture medium (37 °C) for a 20 min incubation at 37 °C in a CO2 incubator.
- Wash cells carefully by dipping the coverslips for 4 s in PBS at RT before fixation in 4% PFA for 20 min. Wash 3 times with PBS and proceed for detection of anti-APP.
- Remove of surface anti-APP antibody bound to non-endocytosed APP by dipping the corresponding coverslips into an acid-stripping buffer (see Recipes) for 5 s and careful washing by dipping the coverslips in PBS at 37 °C for 4 s.
- For assaying the pool of non-recycled APP bound to anti-APP (see Figure 1C), cells are carefully washed for 4 s in PBS at RT, followed by removal of recycled anti-APP antibody by dipping coverslips into an acid-stripping buffer for 5 s before fixation in 4% PFA for 20 min at RT. After washing 3 times with PBS proceed for detection of anti-APP.
Note: Cells are fixed for 20 min instead of 10 min because Neuro2a cells, after the extensive manipulation required to detect the recycled APP, tend to detach from the coverslips.
- Remove cell culture media with a plastic Pasteur pipette, add 500 µl serum-free culture medium at 37 °C and incubate cells for 30 min at 37 °C in cell culture incubator.
- Anti-APP antibody detection (Day 3).
- For detection of the recycled APP bound to anti-APP at the plasma membrane (after 20 min chase, see Figure 1B):
Note: Permeabilization is not necessary since the recycled anti-APP is extracellular.- Place Parafilm stretched on a 24 wells plate lid.
- Place each coverslip (cells facing down) onto a 50 µl droplet of diluted donkey anti-mouse Alexa 488 (1:250) in 3% FBS/PBS.
- Cover the reaction and incubate it for 60 min at RT, in the dark.
- Place coverslips back on a 24-well plate and wash 3 times with PBS at RT.
- Mount coverslips with cells facing down onto a 25 µl droplet of Fluoromount G on microscope slides and let dry overnight in the dark to preserve the fluorescence signal.
- Place Parafilm stretched on a 24 wells plate lid.
- For detecting the pool of non-recycled APP bound to anti-APP (an optional endocytosis control, see Figure 1C)
- Place coverslips on the 24-well plate and add 250 µl of 0.1% saponin/PBS to permeabilize fixed cells for 20 min at RT.
- Wash once with PBS at RT.
- Place Parafilm stretched on a 24 wells plate lid.
- Place each coverslip (cells facing down) onto a 50 µl droplet of diluted donkey anti-mouse Alexa 488 (1:250) in 3% FBS/0.1% saponin/PBS.
- Place coverslips back on a 24-well plate and wash 3 times with PBS at RT.
- Mount coverslips with cells facing down onto a 25 µl droplet of Fluoromount G on microscope slides and let dry overnight in the dark to preserve the fluorescence signal.
- Place coverslips on the 24-well plate and add 250 µl of 0.1% saponin/PBS to permeabilize fixed cells for 20 min at RT.
- For detection of the recycled APP bound to anti-APP at the plasma membrane (after 20 min chase, see Figure 1B):
- Image acquisition (Day 4)
- Image acquisition can be done using epifluorescence microscopy (such as ZEISS Z2) with a 60x NA 1.4 oil immersion objective and a CCD camera (see Equipment for details on microscope used). Exposure times should be determined based on the sample with the brightest expected signal, to use as much of the dynamic range of the camera and a 16-bit binary range as possible, without saturating any of the pixels.
- Acquire 10-20 fields per condition to have sufficient data for statistical analysis. Select fields with isolated cells to facilitate analysis. Focusing on the dorsal plasma membrane may facilitate the visualization of APP recycling, see Figure 2 for representative images.
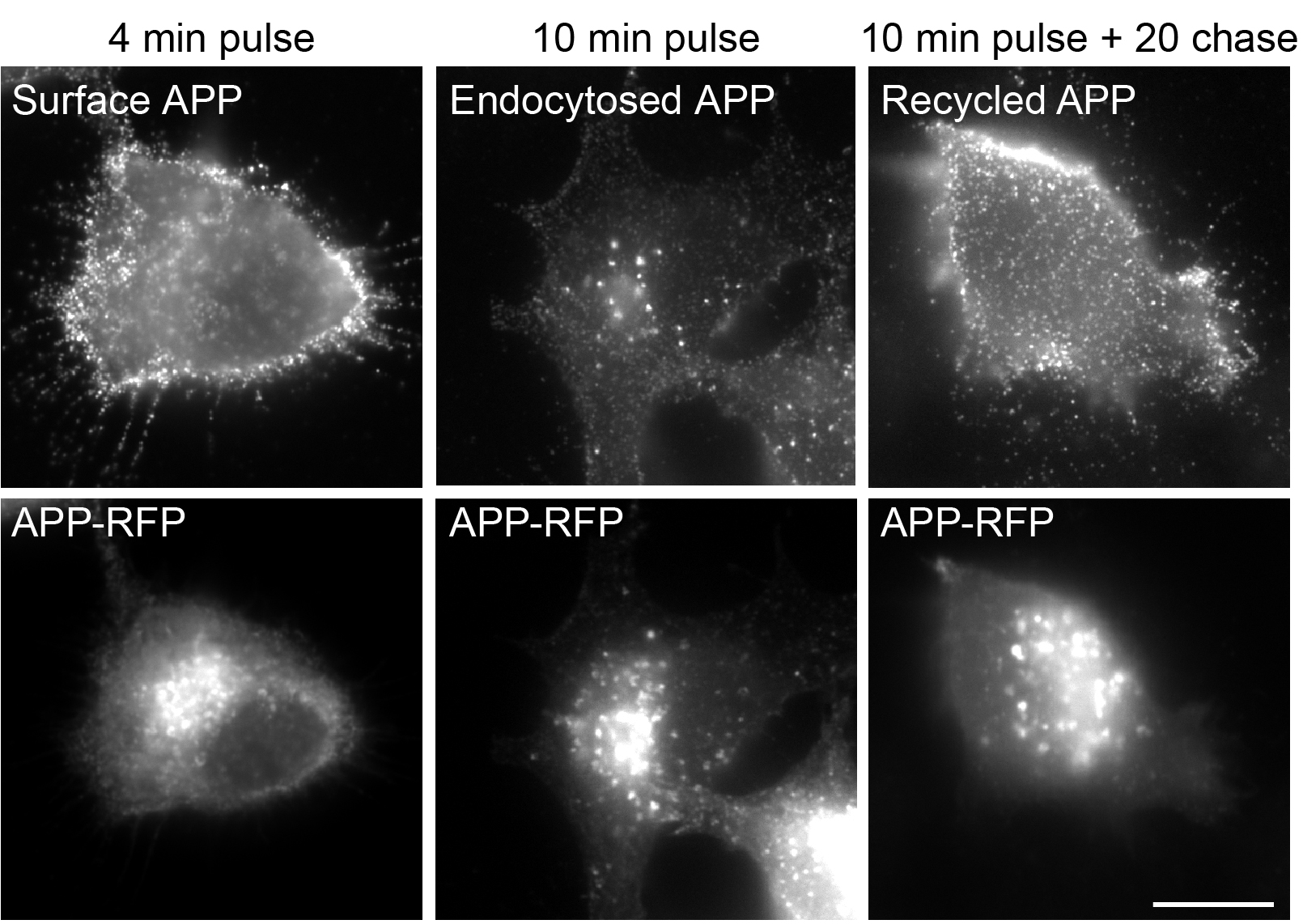
Figure 2. Representative images of APP surface labeling (4 min), endocytosed APP (10 min) and recycled APP (+ 20 min chase) by Neuro2a cells expressing APP-RFP with anti-APP (22C11) detected with anti-mouse Alexa 488 (adapted from Ubelmann et al., 2017). Scale bar = 10 μm.
- Image acquisition can be done using epifluorescence microscopy (such as ZEISS Z2) with a 60x NA 1.4 oil immersion objective and a CCD camera (see Equipment for details on microscope used). Exposure times should be determined based on the sample with the brightest expected signal, to use as much of the dynamic range of the camera and a 16-bit binary range as possible, without saturating any of the pixels.
- Image analysis & APP recycling quantification (Figure 3) (Day 4)
Note: Quantification of fluorescent signal of endocytosed APP per single cell acquired with ImageJ/Fiji.
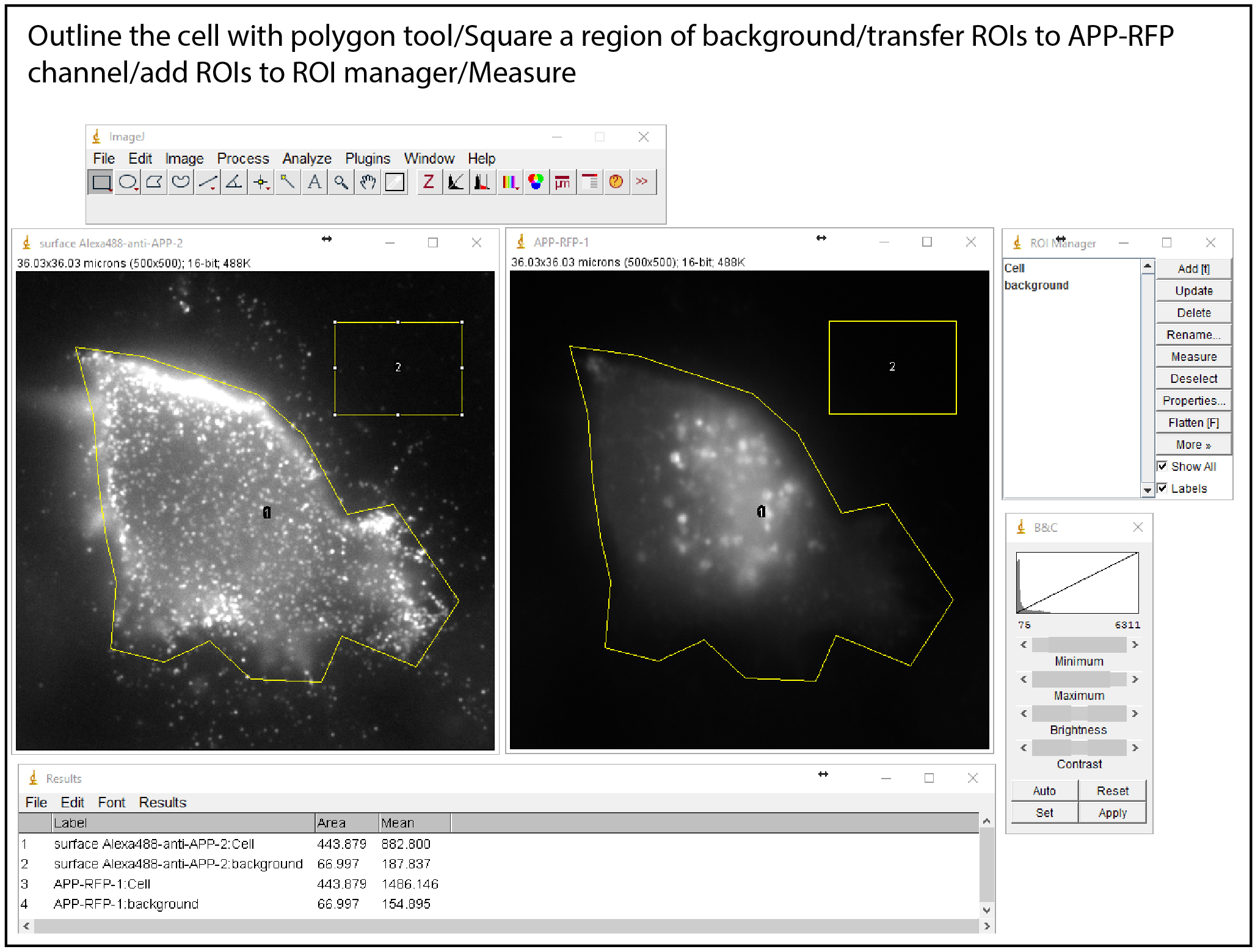
Figure 3. Image analysis of APP recycling
- Select cells with median expression of APP-RFP for image analysis in order to uniformize the quantity of APP present on the surface, available for endocytosis and subsequent recycling. Repeat the procedure for 20-50 cells with good morphology and median APP-RFP expression, about 10 fields per condition.
- Outline each cell based on anti-APP or APP-RFP signal, which becomes visible upon increasing the brightness using B&C tool (without altering intensity levels), using the tool ‘polygon selection’. Add it to ‘ROI manager’ (shift + t).
- Draw a background ROI using the tool ‘polygon selection’ in the original image and add it to the ‘ROI manager’.
- Measure mean intensities of APP-RFP and background ROIs using ‘measure’ (click M) of ROI manager. Fluorescence intensities of the two regions (background and cell) are measured for APP-RFP and anti-APP using the ‘ROI manager’.
- Transfer all regions to recycled Alexa488-anti-APP channel, and use ‘measure’ to obtain the mean fluorescence intensities per ROI and repeat this procedure for each ROI.
- Export measurements to Microsoft Excel.
- Select cells with median expression of APP-RFP for image analysis in order to uniformize the quantity of APP present on the surface, available for endocytosis and subsequent recycling. Repeat the procedure for 20-50 cells with good morphology and median APP-RFP expression, about 10 fields per condition.
Data analysis
Note: Data analysis is done using Microsoft Excel.
- Subtract the background mean fluorescence intensity for all measurements, per single cell.
- Normalize the Alexa488-anti-APP fluorescence intensities by the APP-RFP fluorescence intensity to control for different expression level of APP-RFP, per single cell.
- Sample size is about 20 cells per condition in each independent experiment, based on previous studies. Statistical significance for at least three independent experiments is determined on normal data (D’Agostino-Pearson omnibus normality test) by two-tailed Student’s t-test and for multiple comparisons one-way ANOVA with Tukey’s test using GraphPad Prism 6.
- Statistical significance for nonparametric data is tested by Mann-Whitney test.
- The numerical analysis and graphic representation can be accessed in Ubelmann et al., 2017.
Notes
- In our hands, APP recycling is robust with the average results very reproducible. However, variability among different cells is expected.
- Optimal APP transfection depends on necessary optimal experimental conditions, good quality APP-DNA plasmid (recent midi-prep) and healthy early passage Neuro2a cells.
Recipes
- Acid-stripping buffer
0.5M NaCl
0.2 M acetic acid
- Fixative solution
4% paraformaldehyde (PFA) in PBS, pH 7
Acknowledgments
We thank for the gift of APP-RFP plasmids from Dr. S. Kins (University of Kaiserslautern) and for Neuro2a cells from S. Miserey-Lenkei (Institut Curie). This work has been supported by CEDOC and by a Marie Curie Integration Grant (334366 TrafficInAD FP7-PEOPLE-2012-CIG; Marie Curie Actions, EC), iNOVA4Health–UID/Multi/04462/2013, a program financially supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Educação e Ciência, through national funds and co-funded by FEDER under the PT2020 Partnership Agreement. CGA is Investigator FCT (IF/00998/2012, FCT). FU was the recipient of an FRM postdoctoral fellowship (SPE20130326599) and an FCT post-doctoral fellowship (SFRH/BPD/94186/2013). This protocol was adapted from our recent publication in EMBO reports (Ubelmann et al., 2017). The authors declare no conflict of interest.
References
- Chaufty, J., Sullivan, S. E. and Ho, A. (2012). Intracellular amyloid precursor protein sorting and amyloid-β secretion are regulated by Src-mediated phosphorylation of Mint2. J Neurosci 32(28): 9613-9625.
- Cirrito, J. R., Kang, J. E., Lee, J., Stewart, F. R., Verges, D. K., Silverio, L. M., Bu, G., Mennerick, S. and Holtzman, D. M. (2008). Endocytosis is required for synaptic activity-dependent release of amyloid-β in vivo. Neuron 58(1): 42-51.
- Grbovic, O. M., Mathews, P. M., Jiang, Y., Schmidt, S. D., Dinakar, R., Summers-Terio, N. B., Ceresa, B. P., Nixon, R. A. and Cataldo, A. M. (2003). Rab5-stimulated up-regulation of the endocytic pathway increases intracellular β-cleaved amyloid precursor protein carboxyl-terminal fragment levels and Aβ production. J Biol Chem 278(33): 31261-31268.
- Koo, E. H. and Squazzo, S. L. (1994). Evidence that production and release of amyloid β-protein involves the endocytic pathway. J Biol Chem 269(26): 17386-17389.
- Koo, E. H., Squazzo, S. L., Selkoe, D. J. and Koo, C. H. (1996). Trafficking of cell-surface amyloid β-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci 109 (Pt 5): 991-998.
- Mucke, L. and Selkoe, D. J. (2012). Neurotoxicity of amyloid β-protein: synaptic and network dysfunction. Cold Spring Harb Perspect Med 2(7): a006338.
- Rajendran, L. and Annaert, W. (2012). Membrane trafficking pathways in Alzheimer's disease. Traffic 13(6): 759-770.
- Rajendran, L., Schneider, A., Schlechtingen, G., Weidlich, S., Ries, J., Braxmeier, T., Schwille, P., Schulz, J. B., Schroeder, C., Simons, M., Jennings, G., Knolker, H. J. and Simons, K. (2008). Efficient inhibition of the Alzheimer's disease β-sretase by membrane targeting. Science 320(5875): 520-523.
- Szodorai, A., Kuan, Y. H., Hunzelmann, S., Engel, U., Sakane, A., Sasaki, T., Takai, Y., Kirsch, J., Muller, U., Beyreuther, K., Brady, S., Morfini, G. and Kins, S. (2009). APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci 29(46): 14534-14544.
- Ubelmann, F., Burrinha, T., Salavessa, L., Gomes, R., Ferreira, C., Moreno, N. and Guimas Almeida, C. (2017). Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep 18(1): 102-122.
- Yamazaki, T., Koo, E. H. and Selkoe, D. J. (1996). Trafficking of cell-surface amyloid beta-protein precursor. II. Endocytosis, recycling and lysosomal targeting detected by immunolocalization. J Cell Sci 109 (Pt 5): 999-1008.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ubelmann, F., Burrinha, T. and Guimas Almeida, C. (2017). Measuring the Endocytic Recycling of Amyloid Precursor Protein (APP) in Neuro2a Cells. Bio-protocol 7(23): e2635. DOI: 10.21769/BioProtoc.2635.
Category
Immunology > Immune cell imaging > Epifluorescence Microscopy
Cell Biology > Cell imaging > Fluorescence
Biochemistry > Protein > Immunodetection > Immunostaining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










