- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
GFP-Grb2 Translocation Assay Using High-content Imaging to Screen for Modulators of EGFR-signaling
Published: Vol 7, Iss 17, Sep 5, 2017 DOI: 10.21769/BioProtoc.2528 Views: 8649
Reviewed by: Ralph Bottcherilgen MenderTomas Aparicio

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2336 Views

Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Anastasia Yerushkin [...] Amir Dori
Dec 5, 2025 1224 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
High-content screening is a useful tool to understand complex cellular processes and to identify genes, proteins or small molecule compounds that modulate such pathways. High-content assays monitor the function of a protein or pathway by visualizing a change in an image-based readout, such as a change in the localization of a reporter protein. Examples of this can be the translocation of a fluorescently tagged protein from the cytoplasm to the nucleus or to the plasma membrane. One protein that is known to undergo such translocation is the Growth Factor Receptor-bound protein 2 (GRB2) that is recruited to the plasma membrane upon stimulation of a growth factor receptor and subsequently undergoes internalization. We have used GFP-tagged Grb2 previously to identify genes that are involved in EGFR signaling (Petschnigg et al., 2017). Ultimately, the assay can be adapted to cDNA expression cloning (Freeman et al., 2012) and can be used in early stage drug discovery to identify compounds that modulate or inhibit EGFR signaling and internalization (Antczak and Djaballah, 2016).
Keywords: Grb2Background
Signal transduction by growth factor receptors is essential for cells to maintain proper function and thus requires tight control. Signal transduction by growth factor receptors is initiated by binding of an external ligand (e.g., Epidermal Growth Factor, EGF) to a transmembrane receptor such as the Epidermal Growth Factor Receptor (EGFR) and activation of downstream signaling cascades (Yao et al., 2015). A key regulator of EGFR-signaling is Growth Factor Receptor-bound protein 2 (Grb2), which is composed of an internal SH2 (Src homology 2) domain flanked by two SH3 domains. Grb2 binds to activated growth factor receptors at phosphorylated tyrosine residues through its SH2 domain, thus coupling receptor activation to SOS-Ras-MAPK (Mitogen-activated protein kinase) signaling cascades. The composition of Grb2 suggests that it can dock to a variety of receptors and transduce signals along multiple pathways. Mutations in signaling pathways frequently lead to the development of cancer. Hence, in order to better understand how aberrant signaling can lead to disease, it is important to identify novel signaling molecules in growth factor signaling. To accomplish this, we used the previously established microscopy-based GFP-Grb2 translocation assay that monitors the translocation of cytosolic GFP-tagged Grb2 to subcellular compartments upon expression of a cDNA library (Figure 1). We used this technique to identify novel proteins that can lead to translocation of GFP-Grb2 when overexpressed and in a second stage tested whether these proteins play a role in EGFR-signaling (Petschnigg et al., 2017). Examples that lead to punctate structures include TACC3, a novel EGFR-interactor, and AMPH (Figure 3). TACC3 led to induction of large GFP-Grb2 puncta, whereas AMPH results in formation of multiple small spots (Figure 3), pointing at potentially different mechanisms of those proteins in EGFR-signaling. In our recent study, we further characterized TACC3 and showed that TACC3 specifically binds to oncogenic EGFR variants and showed that TACC3 enhances EGFR-stability at the cell surface and increases EGFR-mediated signaling. The GFP-Grb2 assay can be expanded to multiple more applications. A siRNA/shRNA or CRISPR library could be co-expressed with GFP-Grb2 and translocation subsequently observed following EGF stimulation. As EGF stimulation would sequester GFP-Grb2 to endosomal structures and the plasma membrane, translocation from there upon siRNA/shRNA knockdown or CRISPR knockout could point at possible factors that ablate EGFR-Grb2 interactions and signaling. In a similar way, small molecule compound screens could be done to test for drugs that can specifically disrupt EGFR-Grb2 interactions upon EGF-stimulation (Figure 1). Other options could be to use mutated Grb2-variants that fail to bind to EGFR or other binding partners, thus the assay can give insight into which gene (when overexpressed or knock-downed) or which drug has an influence on specific binding domains of Grb2.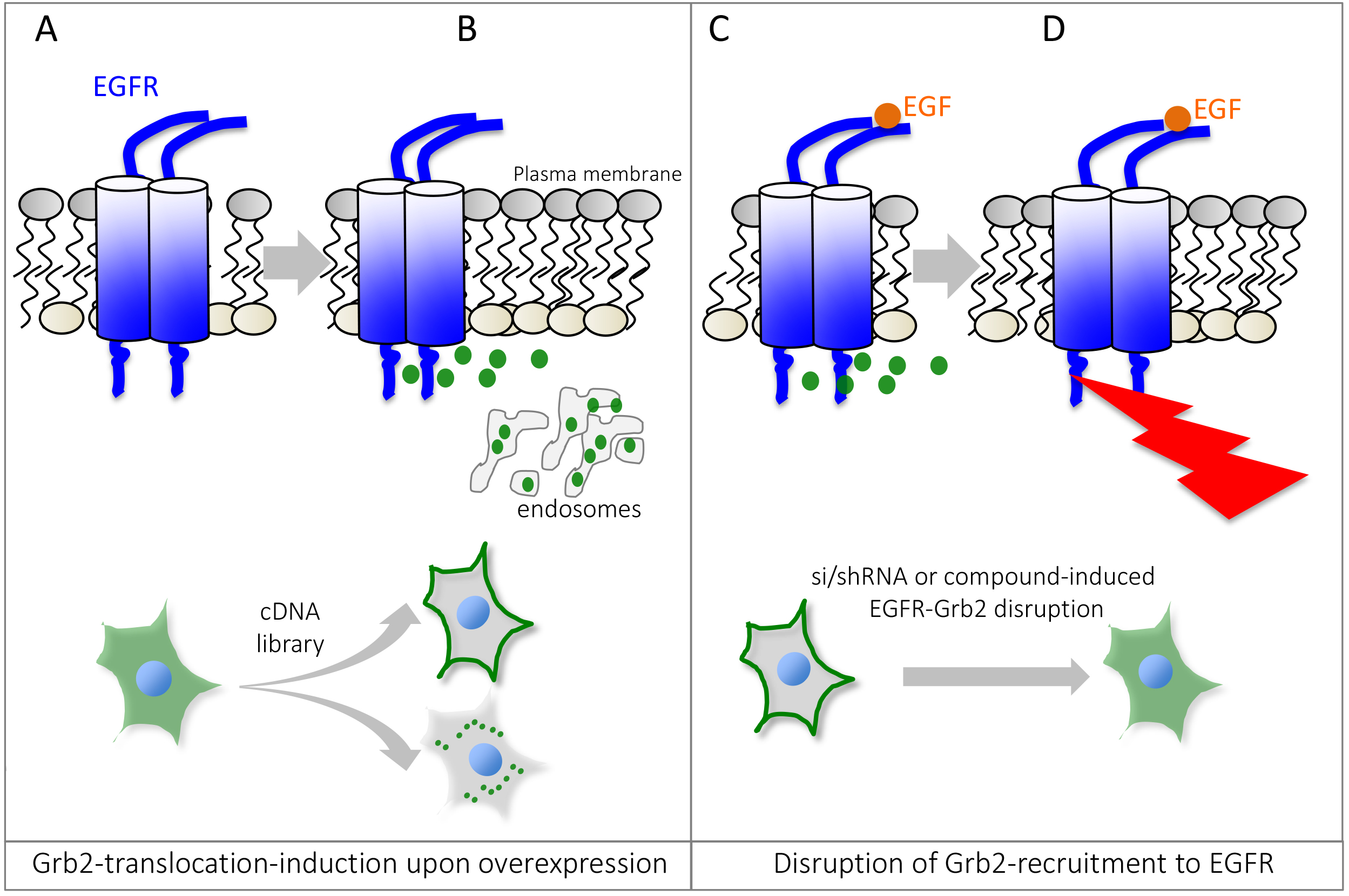
Figure 1. Schematic overview of the Grb2 translocation assay. Under non-stimulated or basal conditions, GFP-tagged Grb2 is mostly found in the cytosol (A), but can be recruited to localized interaction partners such as activated or endocytosed Epidermal Growth Factor Receptor, EGFR. Expression of a cDNA expression plasmid can lead to relocalization of GFP-Grb2 to the plasma membrane, endosomal structures or other sub-cellular locations by binding to Grb2 interaction partners or activation of Grb2-dependent cellular pathways (B). Stimulation with EGF recruits Grb2 to phosphotylated EGFR (C). Small molecule compounds or si/shRNA libraries can help identify genes or drugs that can disrupt Grb2 binding to the EGFR, impairing the recruitment to the receptor and thus predominant cytosolic localization of GFP-Grb2 (D). Since Grb2 can interact with other growth factor receptors as well, the assay can be adapted to monitor interaction/recruitment to other receptors as well.
Materials and Reagents
- Pipette tips (any standard sterile tips can be used, either non-filtered or filtered)
- ViewPlate-96 Black, Optically Clear Bottom (PerkinElmer, catalog number: 6005225 )
- Tissue culture plates: Nunc cell culture Petri dishes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 172931 )
- 0.22 µm filter
- HeLa cells (ATCC, catalog number: CCL-2 ) or HEK293T cells (ATCC, catalog number: CRL-3216 )
- pMOS-GFP-Grb2 plasmid (Ketteler et al., 2002)
- 3xFLAG-TACC3 plasmid (Petschnigg et al., 2017)
- Dulbecco modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 61965026 )
Note: Any commercially available DMEM can be used, and GlutaMax can be added, but is not required for HeLa and HEK293T cells. - Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10500056 )
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Trypsin/EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: R001100 )
- GlutaMax (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 35050061 )
- Paraformaldehyde, 4% (v/v) (Santa Cruz Biotechnologies, catalog number: sc-281692 )
Note: Should be stored at -20 °C in aliquots for long-term storage. Thawed aliquots can be kept at 4 °C for up to a month. - Hoechst 33342 trihydrochloride, trihydrate, 1 mg/ml stock (Thermo Fisher Scientific, InvitrogenTM, catalog number: H3570 )
- Polyethylenimine (PEI), 10 mg/ml stock in water (Sigma-Aldrich, catalog number: 408727 )
- Sodium chloride (NaCl), AnalaR NORMAPUR (VWR, catalog number: 27810.295 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium phosphate dibasic (Na2HPO4), AnalaR NORMAPUR (VWR, catalog number: 102494C )
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P8281 )
- PEI solution (see Recipes)
- 1x phosphate buffered saline (PBS) (pH 7.4) (see Recipes)
- 10x phosphate buffered saline (PBS) (see Recipes)
Equipment
- Multi-channel pipette (Finnpipette, 8-channel P300, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4661030N )
- Incubator (Eppendorf, model: Galaxy® 170 R )
- High-content screening microscope (PerkinElmer, Opera)
Software
- ImageJ
Procedure
- Prepare PEI solution (see Recipes) as 10 mg/ml in water.
Note: Other transfection methods can be used as well, PEI is used here due to being inexpensive and showing good transfection efficiency. - Cultivate HeLa or HEK293T cells in DMEM supplemented with 10% FBS and penicillin-streptomycin at 37 °C and 5% CO2 until 70-80% confluent.
- Trypsinize cells and seed 2 x 104 cells into each well of a PerkinElmer Viewplate using a multi-channel pipette (total volume of 50 µl). Include wells for negative and positive controls (see Note 3). For weakly adherent cells, pre-treatment with gelatin or poly-L-lysine may be required.
- Incubate cells overnight at 37 °C and 5% CO2.
- Prepare a mix of 100 ng GFP-Grb2 plasmid DNA with 100 ng of cDNA (e.g., 3xFLAG-TACC3 as positive control or other controls, see Note 3) in DMEM (25 µl) without FBS and penicillin-streptomycin.
- Add PEI (10 mg/ml solution) at a ratio of 4:1, i.e., for 200 ng total DNA add 0.8 µl PEI.
- Vortex the DMEM/DNA/PEI-mix and leave at room temperature for 15-20 min.
- Add the DMEM/DNA/PEI-mix dropwise to cells using a gentle dispensing method.
Note: Alternatively, the transfection mix can be dispensed first into plates and then cells added on top (reverse transfection). - 6 h after transfection, replace the transfection mix with fresh DMEM plus FBS/penicillin-streptomycin media.
- Incubate cells overnight at 37 °C and 5% CO2.
- The next day, aspirate off the media and wash the cells once with 1x PBS (see Recipes).
Note: At this stage, treatments such as EGF stimulation or drug treatment can be performed before the PBS-wash, depending on the duration of the incubation (e.g., 10 min EGF, or a couple of hours of drugs). - Add 50 µl of 4% PFA in PBS to each well and incubate for 10 min at room temperature.
Note: The assay can also be performed on live cells, in which case steps 12 and 13 can be omitted. Here, we fixed the cells as they can be kept in PBS for a couple of days and plates can be imaged at a later time-point. Yet, live-cell imaging allows for more experimental flexibility, such as time-lapse microscopy. - Aspirate off the PFA and wash once with 1x PBS.
- Add Hoechst 33342 (diluted 1:10,000 in 1x PBS) for 10 min.
- Wash cells once with 1x PBS and add 50 µl 1x PBS.
Note: At this stage, plates can be stored at 4 °C until imaging, up to a couple of days. - Acquire images on a suitable fluorescence microscope (e.g., Opera Phenix) with the following settings: Channel 488 nm for GFP, 100% laser power, 40x objective, binning ‘1’, exposure time may vary between 500 msec and 2 sec, depending on transfection efficiency and brightness. Figure 2 shows the settings on the Opera-high-content screening microscope. Example images (TACC3-overexpression and AMPH-overexpression compared to GFP-Grb2 only) are shown in Figure 3.
- Perform image analysis or qualitatively assess images by eye.
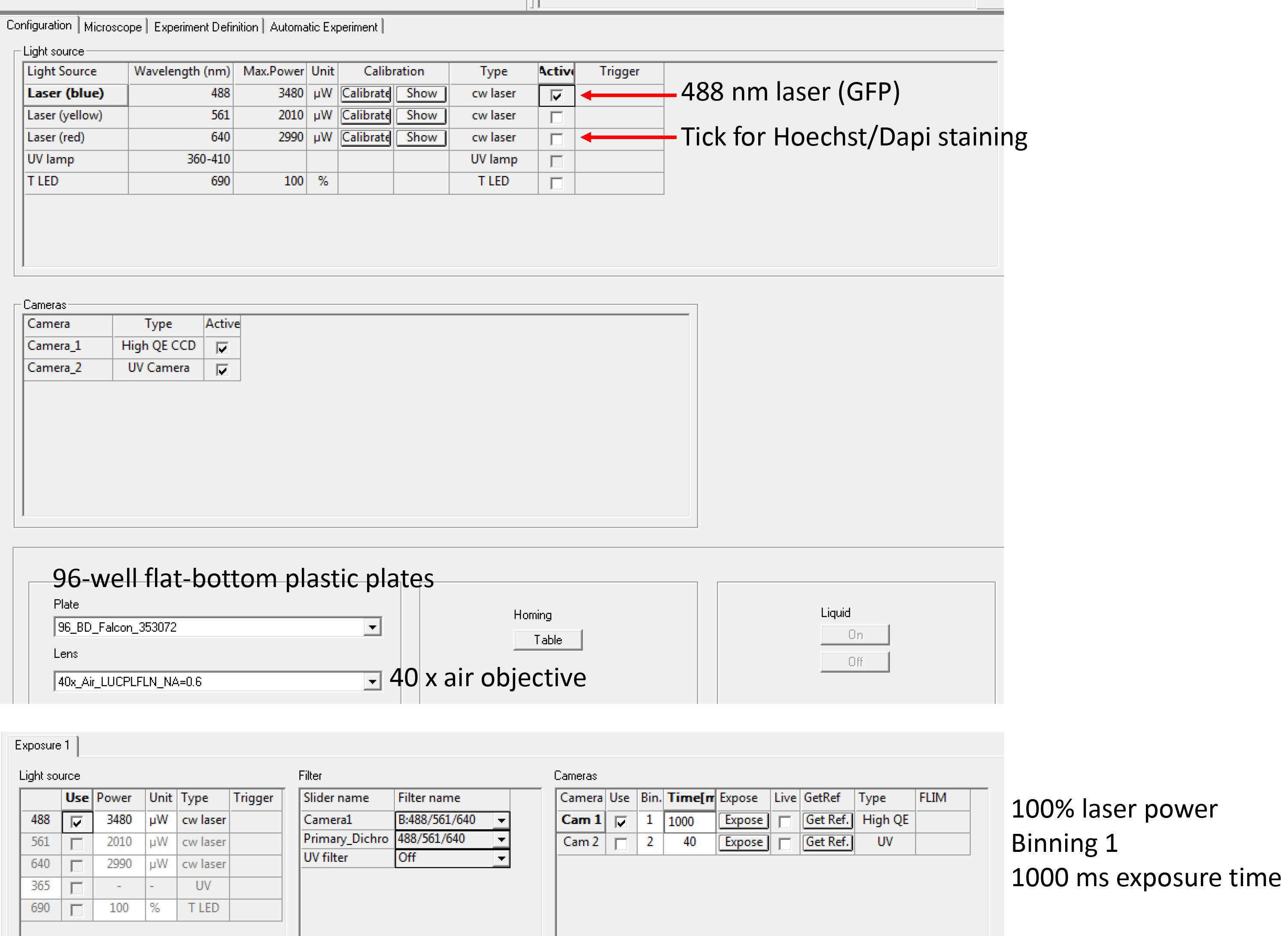
Figure 2. Instrument settings for image acquisition. A screenshot of the Opera LX image acquisition page. Arrows indicate settings that can be adjusted including laser channels, cameras, plate types, objectives, laser power, filters and exposure times (from top to bottom). Other instruments vary in the layout and setup of settings.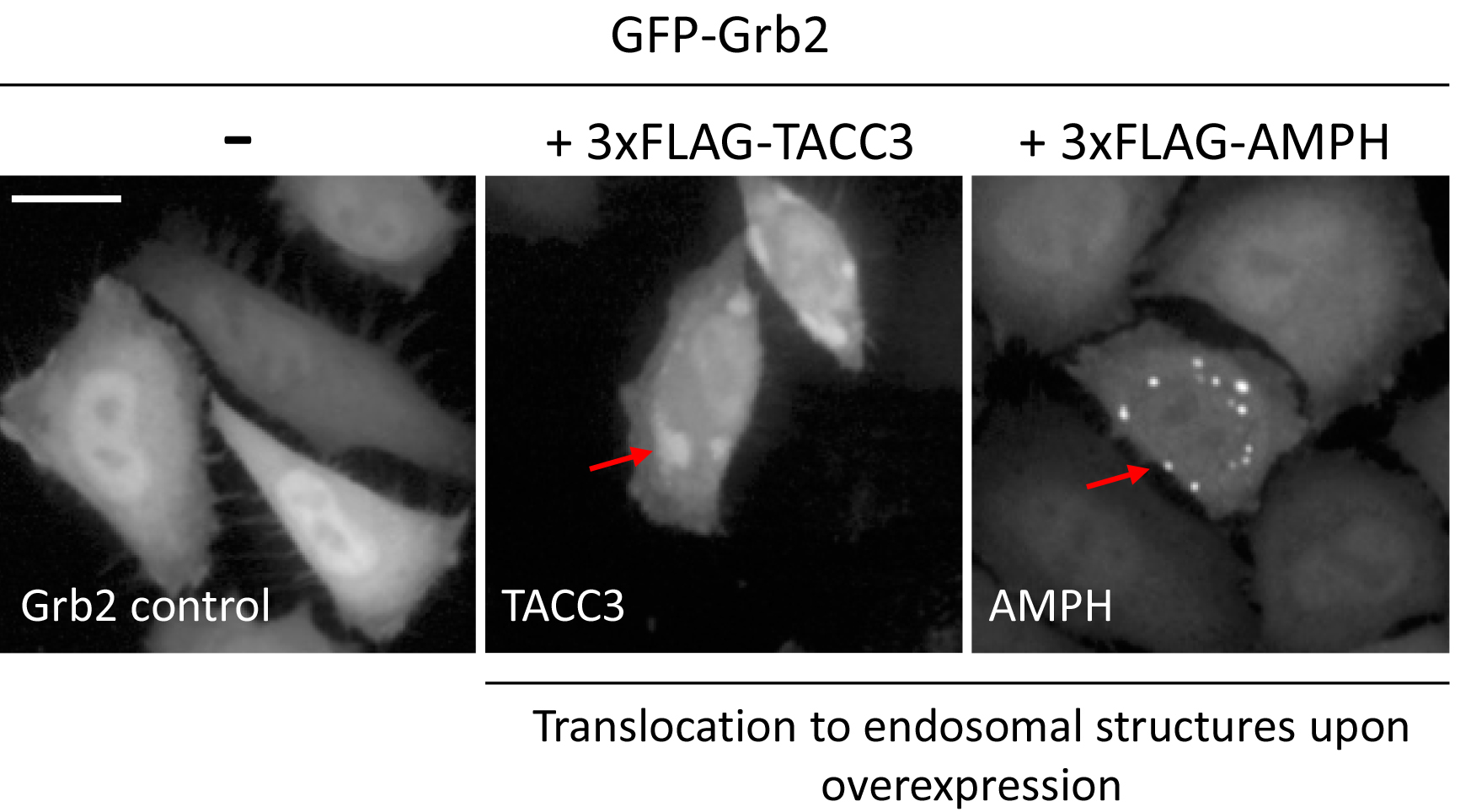
Figure 3. Example images of changes in localization of GFP-Grb2 upon overexpression of TACC3 or AMPH. Arrows indicate punctate structures upon translocation. Scale bar = 20 µm.
Data analysis
Fluorescence images were analysed using the freely available software ImageJ. For our study, structures were analysed individually and different punctate structures were assessed by eye. However, for a high-throughput screen, structures can furthermore be quantified using a custom-made image analysis ImageJ-pipeline. For more details see as an example (Ketteler et al., 2017). Alternatively, other software can be used such as CellProfiler can be used. A typical high-content analysis image processing workflow consists of noise reduction, segmentation, and feature extraction steps. Typically, the original nuclear channel of a sample cell is smoothened with a median filter, segmented with k-means clustering algorithm and the objects are selected and quantified (Ketteler and Kriston-Vizi, 2016).
Notes
- The assay is quite robust and reproducible. However, when performing the assay, it is crucial to ensure that cells are quite low passage (< P30) and regularly passaged (at 70-80% confluency), as this can affect transfection efficiency.
- Transfection efficiency can be variable between plates/wells, hence the generation of stable cell lines, such as cells stably expressing GFP-Grb2, could improve variability. As shown in this protocol, we co-transfected GFP-Grb2 and putative EGFR-interactors such as TACC3 and AMPH. We have also generated stable TACC3-cell lines and transfected GFP-Grb2 and get the same results. Yet, for large-scale screening experiments, we recommend generating GFP-Grb2 stable HeLa or HEK293T cell lines.
- It is important to include controls in the assay to ensure the translocation assay works. In this case, negative control is GFP-Grb2 only (which should appear as cytosolic pattern), and a positive control can be included as well, such as overexpressing TACC3 (3xFLAG-TACC3), which should result in big, punctate GFP-patterns. Other positive controls can be any proteins that are known to induce translocation of Grb2 from the cytosol, such as EGFR, which would sequester Grb2 to the plasma membrane and/or endosomes.
- This protocol describes the use of the translocation assay for screening applications, hence the high-throughput confocal microscope Opera is used. The assay can be performed using any (confocal) fluorescence microscope. Thus, cells can also be seeded onto cover slips or special imaging chambers. The protocol is substantially the same after seeding.
- The assay can be adapted to many more applications as stated in the background section. Time of drug incubations, EGF-stimulation or siRNA/shRNA expression needs to be determined individually by each user.
- Both HeLa and HEK293T cells have been successfully used for our assay. Yet, it should be noted that HeLa cells are easier to work within 96-well plates as they do not tend to dislodge from the plates during the washing steps (before the 4% paraformaldehyde fixing). If using low-adherent cells, plates can be pretreated with gelatin or poly-L-lysine. The assay can also be done in live cells, in which case the user can omit the fixing steps.
Recipes
- PEI solution
Prepare a 10 mg/ml solution in sterile distilled water and filter-sterilize (0.22 µm filter) and store at 4 °C
Note: Prepare fresh solution about once a month to ensure equal transfection efficiency. - 1x phosphate buffered saline (PBS) (pH 7.4)
137 mM NaCl
10 mM phosphate
2.7 mM KCl
Note: Prepare a 10x PBS stock and dilute with sterile water to 1x PBS. - 10x phosphate buffered saline (PBS)
80 g NaCl
2 g KCl
14.4 g Na2HPO4
2.4 g KH2PO4
Dissolved in 800 ml distilled H2O
Adjust pH to 7.4
Fill up to 1 L
Acknowledgments
This work was supported by the UK Medical Research Council core funding to the MRC-UCL University Unit (MC_EX_G0800785) and a BBSRC New Investigator Award to RK (BB/JO/5881/1). JP was supported by an EC-Marie Curie International Incoming Fellowship (FP7-PEOPLE-2013-IIF). This protocol has been adapted from (Petschnigg et al., 2017).
References
- Antczak, C. and Djaballah, H. (2016). A High-content assay to screen for modulators of EGFR function. Methods Mol Biol 1360: 97-106.
- Freeman, J., Kriston-Vizi, J., Seed, B. and Ketteler, R. (2012). A high-content imaging workflow to study Grb2 signaling complexes by expression cloning. J Vis Exp (68).
- Ketteler, R., Freeman, J., Ferraro, F., Bata, N., Cutler, D. F. and Kriston-Vizi, J. (2017). Image-based siRNA screen to identify kinases regulating Weibel-Palade body size control using electroporation. Sci Data 4: 170022.
- Ketteler, R., Glaser, S., Sandra, O., Martens, U. M. and Klingmuller, U. (2002). Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther 9(8): 477-487.
- Ketteler, R. and Kriston-Vizi, J. (2016). High-content screening in cell biology. In: Bradshaw, A. R. and Stahl, D. P. (Eds). Encyclopedia of Cell Biology, Vol 4. Academic Press pp: 234-244.
- Petschnigg, J., Kotlyar, M., Blair, L., Jurisica, I., Stagljar, I. and Ketteler, R. (2017). Systematic identification of oncogenic EGFR interaction partners. J Mol Biol 429(2): 280-294.
- Yao, Z., Petschnigg, J., Ketteler, R. and Stagljar, I. (2015). Application guide for omics approaches to cell signaling. Nat Chem Biol 11(6): 387-397.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Petschnigg, J. and Ketteler, R. (2017). GFP-Grb2 Translocation Assay Using High-content Imaging to Screen for Modulators of EGFR-signaling. Bio-protocol 7(17): e2528. DOI: 10.21769/BioProtoc.2528.
Category
Cancer Biology > Cancer biochemistry > Protein
Cell Biology > Cell staining > Protein
Cell Biology > Cell signaling > Intracellular Signaling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








