- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Superoxide Dismutase (SOD) and Catalase (CAT) Activity Assay Protocols for Caenorhabditis elegans
Published: Vol 7, Iss 16, Aug 20, 2017 DOI: 10.21769/BioProtoc.2505 Views: 38796
Reviewed by: Peichuan ZhangKristin ShinglerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1758 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1575 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views
Abstract
Assays for superoxide dismutase (SOD) and catalase (CAT) activities are widely employed to indicate antioxidant responses underlying the toxic effects of test chemicals. Yet, earlier studies mainly described the procedures as performed according to manufacturer’s instructions without modifications that are specific to any organisms. The present protocol describes the steps in analyzing the superoxide dismutase (SOD) and catalase (CAT) activities in C. elegans, which is a model organism that can be used to study effects of pharmaceutical compounds and environmental pollutants. The main steps include: (1) sample preparation; (2) total protein assay; (3) SOD activity assay; (4) CAT activity assay; and (5) medium list and formula, and also data analysis and performance notes.
Keywords: SODBackground
Biomarkers are essential to examine biological and pathogenic processes in response to a chemical, an agent or a therapeutic intervention. Various biological processes in organisms result in reactive oxygen species (ROS) which cause oxidative stress. In response to such oxidative stress, organisms can deploy superoxide dismutase (SOD) and catalase (CAT) to scavenge ROS so as to protect the cellular homeostasis (Balaban et al., 2005). On the one hand, various chemicals (pollutants) can retard such antioxidant responses, and disturb the health of organisms including human beings. On the other hand, many pharmaceuticals aim to strengthen the antioxidant responses to improve health. Therefore, activities of SOD and CAT are very important to reflect potential effects of chemicals or/and pharmaceuticals.
Caenorhabditis elegans (C. elegans) is a model organism that has been used to study effects of pharmaceutical compounds (Dengg and van Meel, 2004; Carretero et al., 2017) and environmental pollutants (Yu et al., 2013a and 2017). Several studies have used SOD and CAT assays to indicate the antioxidant responses and potential mechanism underlying the toxic effects of test chemicals (Feng et al., 2015; Yu et al., 2012; 2016 and 2017). However, these studies simply described that the determination was carried out, by using a generic kit protocol without species-specific modifications. Therefore, the explicit protocols to perform SOD and CAT assays in C. elegans are still needed for better specific instruction.
In the present protocol, we provide a nematode protocol with experimental details to analyze SOD and CAT activities in C. elegans.
Materials and Reagents
- Pipette tips (https://online-shop.eppendorf.com)
- Plate (60 mm)
- Centrifuge tubes, 1.5 ml (Eppendorf, catalog number: 022364111 )
- Absorbent paper (KCWW, Kimberly-Clark, catalog number: 0131 )
- 96-well plate, with lids (Corning, Costar®, catalog number: 3599 )
- Sealing tape, for 96-well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 15036 )
- Nematodes (wild type N2)
Note: These nematodes are treated according to each researcher’s experiments. In the present protocol, the nematodes only have difference in numbers. - BCA protein assay kits (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23225 or 23227 )
- Protein standard (5 mg/ml)
- BCA reagent A and B
- Protein standard (5 mg/ml)
- SOD assay kits (Beyotime Biotechnology, catalog number: S0101 )
- Reaction initiation solution
- SOD detection buffer
- WST-8
- Enzyme solution
- Reaction initiation solution (40x)
- Reaction initiation solution
- CAT assay kits (Beyotime Biotechnology, catalog number: S0051 )
- Enzyme conjugate
- Wash solution 40x
- Substrate A and B
- Stop solution
- Enzyme conjugate
- Sodium hydrate (NaOH), Analytic Reagent (Sinopharm Chemical Reagent, catalog number: 10019762 )
- Sodium hypochlorite (Antiformin, NaOCl, with 6-14% active Cl), Analytic Reagent (ALADDIN, catalog number: S101636-500 ml )
- Potassium phosphate dibasic (K2HPO4), Analytic Reagent (Sinopharm Chemical Reagent, catalog number: 20032118 )
- Potassium phosphate monobasic (KH2PO4), Analytic Reagent (Sinopharm Chemical Reagent, catalog number: 10017618 )
- Clorox solution (see Recipes)
- Phosphate buffered saline/buffer (PBS), pH 7.0 (see Recipes)
Equipment
- Pipettes
- Microscope
- Centrifuge, Eppendorf 5417R (Eppendorf, model: 5417 R , catalog number: 01396)
- Pestles, Eppendorf (Eppendorf, catalog number: F0140010 )
- Motor-driven tissue grinder (Beijing Baiwan Electronic Technology, catalog number: HG215-LH-A )
- Ice bath, in centrifuge tube box
- Microplate reader, BioTek (BioTek Instruments, model: Epoch )
- Sterilized bottle, Fisherbrand (100 ml, Fisher Scientific, catalog number: FB800100 ; 250 ml, Fisher Scientific, catalog number: FB800250 ; 500 ml, Fisher Scientific, catalog number: FB800500 )
- Magnetic stir bar
- Magnetic stir plate
- pH meter
- Incubator, Yiheng (Yiheng, model: LRH-1000F )
Procedure
- Sample preparation
- Sample collection
The entire protocol uses the L4 stage nematodes as the reference point. Follow the nematode culture and age-synchronization according to earlier reports (Solis and Petrascheck, 2015; Yu et al., 2017). Wash the L4 stage nematodes off each nematode growth medium plate (60 mm) with 1.6 ml phosphate buffered saline (PBS, pH 7.0) into a 1.5 ml centrifuge tube. After a 30-min settlement by gravity at 4 °C, carefully pipette out and discard the supernatants. Add 500 μl ice-cold PBS, resuspend the pellets with gentle and repetitive pipetting followed by a further 15-min settlement. Then, discard the supernatants, add 500 μl ice-cold PBS, resuspend the pellets with gentle and repetitive pipetting and obtain groups with 20, 50, 100, 200, 300, 400 and 500 nematodes in 200 μl PBS in the centrifuge tubes using the following steps.
Take the group with 100 nematodes as an example.- Count the nematodes in 20 μl PBS under a microscope with three independent trials to estimate the nematode density in the whole tube.
- Based on the number of worms obtained, calculate and adjust the volume of PBS so that the final density is ~9 to 11 worms per 20 μl. For example, if the number is more than 10, add more PBS accordingly; and if the number is less than 10, perform a 10-min settlement and remove corresponding amount of PBS to achieve the desired density.
- Transfer 260 μl PBS with the nematodes into new 1.5 ml tubes with pipette, and resuspend the pellets with gentle and repetitive pipetting.
- Count nematode numbers in 20 μl PBS in three independent trials, with gentle and repetitive pipetting in between, to confirm the nematode density, leaving 200 μl in the tube. If the nematode density is not consistent with the setting, repeat step A1b.
- Prepare groups with other nematode numbers through the same way.
- Count the nematodes in 20 μl PBS under a microscope with three independent trials to estimate the nematode density in the whole tube.
- Sample storage
Centrifuge the tubes at 5,000 x g for 5 min (4 °C). Discard the supernatant PBS carefully with pipette and store the pellets at -26 °C overnight or -80 °C for over a week. - Sample homogenization
- Connect a clean pestle with the motor-driven tissue grinder. Wash the pestle in ice-cold PBS, and clean it with the absorbent paper. Push the pestle tightly against the pellets in the 1.5 ml centrifuge tube. Keep the centrifuge tube in an ice bucket (see Figure 1). Start the tissue grinder, grind the pellets for 10 sec with a 2-sec cooling down followed by another 10-sec grinding. Then, use 200 μl ice-cold PBS to wash the residual liquids on the pestle back to the centrifuge tube (see Figure 1), before taking it out of the tube.
- Centrifuge the tubes at 5,000 x g for 5 min at 4 °C. Pipette out 200 μl supernatants from each sample. Aliquot them into four tubes with 50 μl in each for subsequent determination.
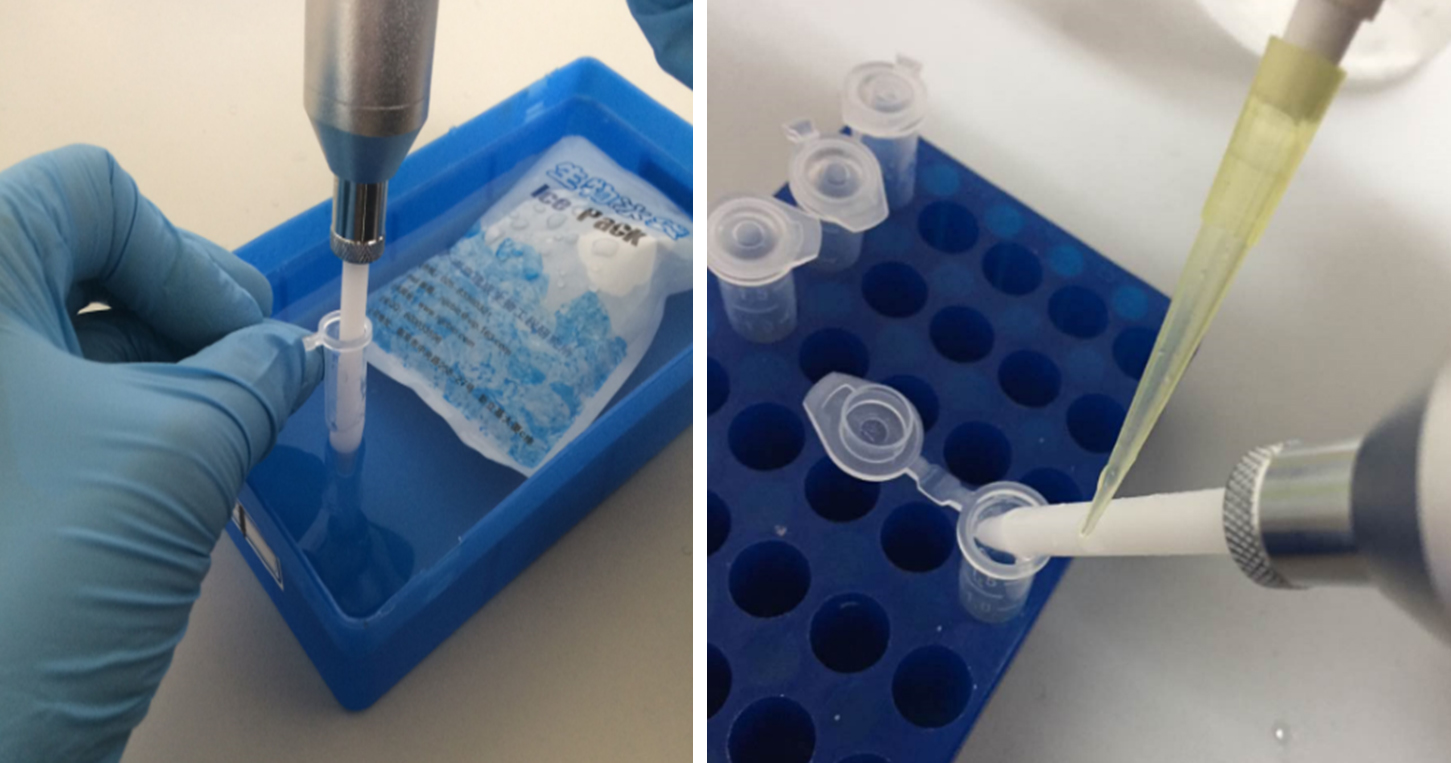
Figure 1. Sample homogenization. The left picture shows that motor-driven tissue grinder homogenizes the pellets in the centrifuge tube in an ice bath, and the right one shows that residual liquids are washed off the pestle with 200 μl ice-cold PBS.
- Connect a clean pestle with the motor-driven tissue grinder. Wash the pestle in ice-cold PBS, and clean it with the absorbent paper. Push the pestle tightly against the pellets in the 1.5 ml centrifuge tube. Keep the centrifuge tube in an ice bucket (see Figure 1). Start the tissue grinder, grind the pellets for 10 sec with a 2-sec cooling down followed by another 10-sec grinding. Then, use 200 μl ice-cold PBS to wash the residual liquids on the pestle back to the centrifuge tube (see Figure 1), before taking it out of the tube.
- Sample collection
- Total protein assay
Measure the total protein (TP) in each sample by enzyme-linked immunosorbent assay (ELISA) kits based on BCA methods.- Preparation for protein standards
Use the stock protein standard (5 mg/ml) to prepare 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2 mg/ml dilutions, 100 μl each using PBS. - Preparation for BCA working solution
Prepare BCA working solution with BCA reagent A and B by a volume ratio of 50:1. Total volume of the BCA working solution is calculated according to the amount of tested samples (200 μl per sample). - Determination of protein concentration
- For each protein standard, add 20 μl into a 96-well plate with at least two replicates. Use 20 μl PBS buffer as control.
- For each nematode sample, add 20 μl from one aliquot into the 96-well plate with at least two replicates. Use up this aliquot to ensure replicate and avoid repetitive freezing-unfreezing of the samples.
- Then, add 200 μl BCA working solution per well for all the protein standards and nematode samples. Seal the wells by sealing tape to avoid the evaporation of water, and incubate the plate for 30 min at 37 °C.
- Measure the absorbance at 562 nm (A562) using a microplate reader.
- For each protein standard, add 20 μl into a 96-well plate with at least two replicates. Use 20 μl PBS buffer as control.
- Data calculation
Calculate the protein contents in the samples according to the standard curve.
One presentation of the relation between the nematode numbers and the protein concentrations is shown in Figure 2.
Figure 2. The relationship between the nematode numbers and the nematode protein concentrations
- Preparation for protein standards
- SOD activity assay
Measure the SOD in each sample by ELISA kits based on WST-8 method. The WST-8 method is based on the colorimetric reaction of WST-8. In the reaction, xanthine oxidase (XO) catalyzes the oxidation conversion of xanthine to yield superoxide anion, which quenches WST-8 to produce water-soluble formazan (a purple dye). As SOD quenches superoxide anion, thus its activity inhibits the overall colorimetric reaction (see Figure 3). Therefore, the inhibition levels are used to indicate the SOD activities in cells, tissues or other biological samples.
Figure 3. The colorimetric reaction of WST-8 with the superoxide anion from xanthine, and the SOD activities inhibit the colorimetric reaction- Preparation of necessary solution
Prepare the standards and controls, and working solutions according to the manufacturer’s instruction of the ELISA kit.- Each sample needs 160 μl WST-8/enzyme working solution and 20 μl reaction initiation solution. Calculate the total amount according to the number of nematode samples plus standards. For WST-8/enzyme working solution, 160 μl consists of 151 μl SOD detection buffer, 8 μl WST-8 and 1 μl enzyme solution.
- Prepare the reaction initiation solution, by diluting from the original stock 1:40 in SOD detection buffer (i.e., 1 μl original solution to 39 μl buffer).
- Each sample needs 160 μl WST-8/enzyme working solution and 20 μl reaction initiation solution. Calculate the total amount according to the number of nematode samples plus standards. For WST-8/enzyme working solution, 160 μl consists of 151 μl SOD detection buffer, 8 μl WST-8 and 1 μl enzyme solution.
- Determination of samples
- Add 20 μl standards and controls into a 96-well plate with at least two replicates.
- Add 20 μl nematode samples into the 96-well plate with at least two replicates. Notably, each nematode sample is processed and divided into 4 aliquots after the step of sample homogenization. Use up one aliquot to ensure replicate and avoid repetitive freezing-unfreezing of the samples.
- Use a multi-channel pipette to add 160 μl WST-8/enzyme working solution into each sample, and then, add 20 μl reaction initiation working solution. The concentrations of standards are 100, 50, 10, 5, 2.5, 1.25 and 0.625 U/ml.
- There are two blank controls. For the blank control 1, 20 μl sample are replaced by SOD detection buffer. For the blank control 2, 20 μl sample and 20 μl reaction initiation solution are replaced by 40 μl SOD detection buffer.
- Incubate the plate for 30 min at 37 °C.
- Read the absorbance at 450 nm (A450) by a microplate reader.
- Add 20 μl standards and controls into a 96-well plate with at least two replicates.
- Calculation in SOD activity assay
- Inhibition (%) = (ABlank control 1 - AStandard or Sample)/(ABlank control 1 - ABlank control 2) x 100%. Use the standards with known SOD activities and their inhibitions to establish a standard curve. Then, use the curve to calculate the SOD activities of samples by their inhibitions. One example for the standard curve is shown in Figure 4.
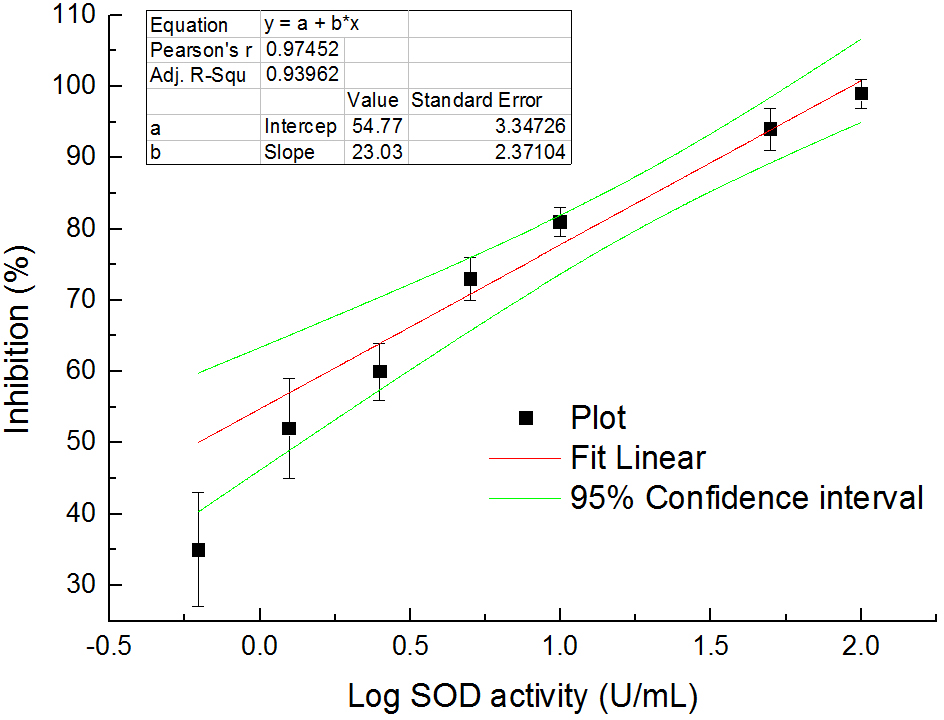
Figure 4. The relationship between SOD activities in logarithm and inhibition (%) on WST-8 colorimetric reaction - Express the SOD enzyme activity in each nematode sample as its proportion (P) in the total protein (TP) of the same sample to eliminate the differences of nematode numbers among samples. The normalized SOD activities in the nematodes are listed in Table 1. The proportion values generally range from 4.04 to 4.35. The values in groups with 20 and 50 nematodes are quite greater than the general range. Although these values are within mean value + 3 standard deviation (SD), we recommend not using them to avoid unreliability. These results indicate that at least 100 nematodes should be used to ensure the feasibility and stability in measuring SOD activities.
Table 1. Connection between normalized SOD activities and nematode numbers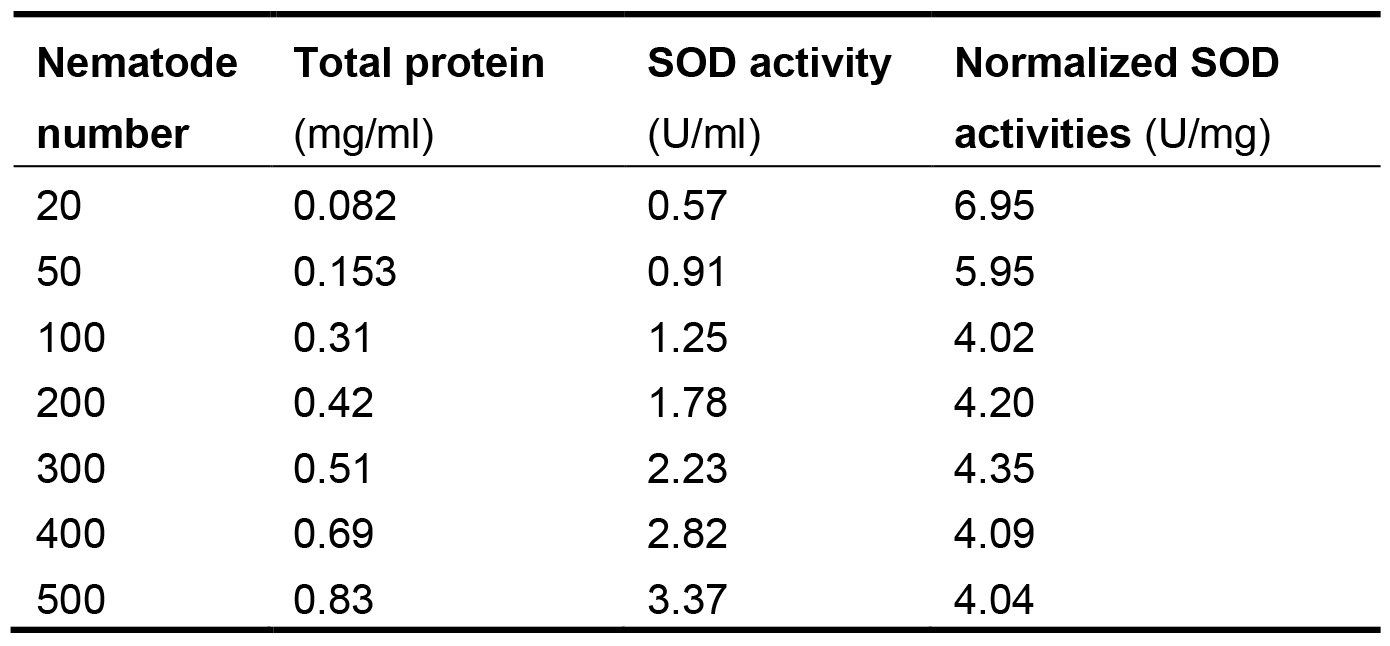
- Inhibition (%) = (ABlank control 1 - AStandard or Sample)/(ABlank control 1 - ABlank control 2) x 100%. Use the standards with known SOD activities and their inhibitions to establish a standard curve. Then, use the curve to calculate the SOD activities of samples by their inhibitions. One example for the standard curve is shown in Figure 4.
- Preparation of necessary solution
- CAT activity assay
Measure the CAT activities in each sample with ELISA kits. The assay principle is based on the reaction of catalase to decompose hydrogen peroxide (H2O2). The excess H2O2 can form complexation with ammonium molybdate to produce light yellow solution. The solution’s adsorption at 450 nm can be used to calculate the concentration of H2O2 and therefore to indirectly indicate the CAT activities.- Reagents preparation
According to the manufacturer’s instruction of the ELISA kit, prepare all reagents before the assay with careful calculation based on the numbers of samples, standards and controls. - Addition of standards or samples
Add 50 μl standards or samples into a 96-well plate. Use 50 μl PBS as blank controls. All standards, samples and controls are recommended to have at least two replicates. Notably, each nematode sample is processed and divided into 4 aliquots after the step of sample homogenization. Use up one aliquot to ensure replicate and avoid repetitive freezing-unfreezing of the samples. The concentrations of standards are 400, 100, 25, 6.25, 1.5625, 0.4 and 0.1 U/L. - Enzyme conjugation
Add 100 μl of enzyme conjugate to wells containing standards, samples and blanks. Then, seal the wells by sealing tape to avoid the evaporation of water, and incubate the microplate statically for 60 min at 37 °C. - Wash
- Discard the liquid content and tap the assay plate on a piece of absorbent paper to remove residual buffer.
- Add 350 μl wash solution (1x, which was diluted from the 40x package with PBS) to each well and keep the microplate static for 1 min. Then, discard the liquid content and tap the assay plate on a piece of absorbent paper to remove residual buffer.
- Repeat the above wash process for four times.
- Discard the liquid content and tap the assay plate on a piece of absorbent paper to remove residual buffer.
- Addition of substrates
Use a multi-channel pipette to add 50 μl substrate A and 50 μl substrate B to each well. Incubate the microplate at 37 °C in the dark for 15 min. - Endpoint measurement
Add 50 μl stop solution to each well. After 1 min incubation, measure the absorbance at 450 nm on a microplate reader within 15 min. - Data calculation and presentation
Calculate the total CAT activities of samples according to the standard curve (see Figure 5), the method of which is similar to that used by the SOD activity assay.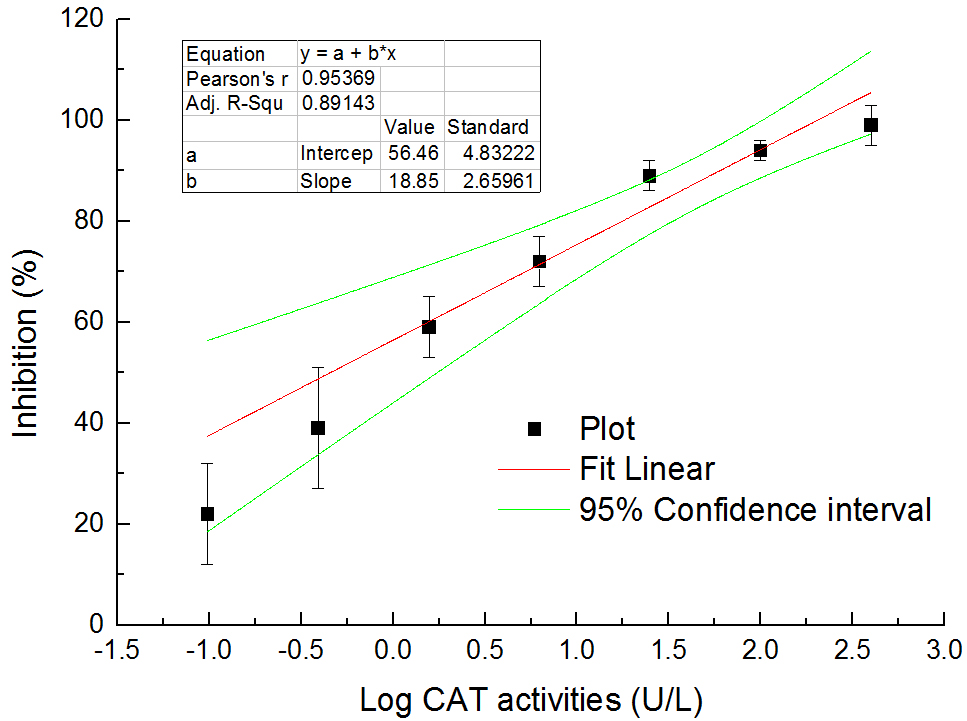
Figure 5. The relationship between total CAT activities in logarithm and inhibition (%) on colorimetric reaction between H2O2 and ammonium molybdate
Express the CAT activities in each nematode sample as its proportion (P) in the total protein (TP) of the same sample to eliminate the differences of nematode numbers among samples. The normalized CAT activities in the nematodes are listed in Table 2. The proportion values generally range from 8.74 to 10.36. The values in groups with 20 and 50 nematodes are significantly greater than this range. These results indicate that at least 100 nematodes should be used to ensure the feasibility and stability in measuring CAT activities.
Table 2. Connection between normalized CAT activities and nematode numbers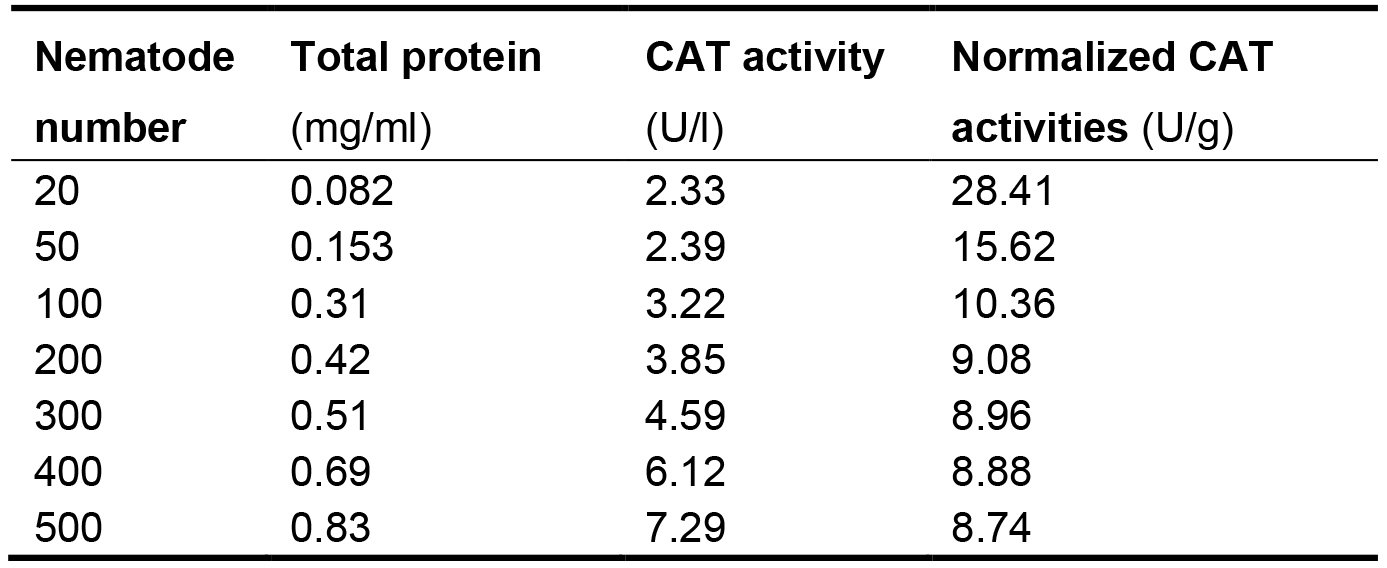
- Reagents preparation
Data analysis
- The data analysis procedure refers to our previous report (Yu et al., 2013b). Firstly, the values of the nematode total protein (TP) in one aliquot of each sample are measured in at least three replicates, and the means of these TP values (TP mean) are calculated to represent the TP concentration in the sample. Secondly, the SOD (or CAT) values in other aliquots of the same sample are also measured in at least three replicates. Thirdly, the SOD (or CAT) values in each replicate are calculated as their proportions to the TP mean value to obtain SOD (or CAT) proportion values. Fourthly, the SOD (or CAT) proportion values are calculated to obtain the mean SOD (or CAT) proportion in the sample and corresponding standard error.
- In cases that the samples come from different treatments, the mean SOD (or CAT) proportion in samples from the control are normalized to represent 100% (or 1.0); then, the SOD (or CAT) proportions in samples from the chemical exposure are transformed to percentages of the controls (POCs) (or fold changes against control); next, the POC values (or fold-changes) in the samples are used to calculate the mean POC and corresponding standard error.
- The nematode samples should be collected from at least three independently repeated experiments, with one set of data in each experiment. The ANOVA (Origin 9.0, Origin Lab Corp., USA) was carried out between the mean SOD (or CAT) proportions in the control and those in the chemical exposure from these independently repeated experiments. The probability levels of 0.05 were considered statistically significant (P < 0.05).
Notes
- When collecting the nematodes from nematode growth medium plates, the bacteria in the worm slurry would cause problems to the assays. Therefore, don’t use centrifugation, but use gravity settlement for sample collection. To fully get rid of the bacteria, gently wash the pellets (i.e., resuspend-settlement-discard supernatant) several times.
- Remember to avoid unnecessary stress on the nematodes. On one hand, use ice-cold PBS to lessen the stress on worms. On the other hand, it is better to spend less time on transfer and other performance with well-trained handling. Use a multi-channel pipette to make simultaneous addition of all solutions, so as to shorten the differences of reaction time among samples. Also, keep all the samples with the same performance to ensure the comparability between samples.
- Considering that the worms will stick to the tip and they tend to settle with gravity, resuspend the pellets with gentle and repetitive pipetting every time when transferring the worms. In this way, we can ensure that the worm density is uniform in the tube, and also that the worms stuck to the tip reach the equilibrium without further sticking.
- To ensure reproducibility and variability, there should be at least 100 nematodes in each tube.
- The supernatants from the homogenized PBS solutions of 100 nematodes are enough for four aliquots, which are adequate for four biochemical assays including the total protein. If more biochemical assays are performed simultaneously, the nematode numbers should increase.
- The protein concentration range of the protein standard solutions can be altered according to preliminary experiments, especially in cases where the nematode numbers are not counted.
- When homogenizing nematode samples, handle the pestles in a uniform manner among different samples. Keep the movement in the same strength with same time duration (10 sec homogenizing + 2 sec cooling down + 10 sec homogenizing for each tube), so as to improve the reproducibility of the data. Also, clean the pestle before using them for another tube, and keep the clean procedure uniform throughout the whole homogenizing procedure.
Put pestles (for subsequent homogenization) in ice-cold PBS. After each use, wash the pestle in ice cold water, and put it back to the clean ice-cold PBS. This performance will ensure the cleanness of the pestle and also avoid potential influence of room-temperature pestles on protein stability of the samples. - If the nematode number in each tube is much more than 500, the duration of homogenizing procedure should be extended to at least twice the normal time (e.g., 10 sec grinding + 2 sec cooling + 10 sec grinding + 2 sec cooling + 10 sec grinding + 2 sec cooling +10 sec grinding). The pestles should be washed with 400 μl PBS. Through this way, the homogenizing efficiency can be ensured, and the total protein concentration in the supernatants can be diluted to better perform the biochemical assays.
- In the prevention of water evaporation, a humid chamber will help.
Recipes
- Clorox solution
Take 30 ml as an example. It consists of 0.6 g NaOH, 5 ml NaOCl (with 6-14% active Cl), and 25 ml deionized H2O, and the final concentrations are 0.5 M and ~1% for NaOH and NaOCl, respectively - Phosphate buffered saline/buffer (PBS), pH 7.0
If a ready-mix is not available, then make the solution as follows:- Solution A: dissolve 17.418 g of K2HPO4 (FW = 174.18) in 1,000 ml in sterilized deionized water in sterilized bottle
- Solution B: dissolve 13.689 g of KH2PO4 (FW = 136.89) in 1,000 ml in sterilized deionized water in sterilized bottle
- In a 2,000 ml flask with a magnetic stir bar on a magnetic stir plate, add 750 ml solution B. After turning on the magnetic stirring apparatus, carefully put the probe of the pH meter underneath the water surface and above the magnetic stirrer. Then, add solution A into the flask slowly, and monitor the pH changes at the same time until the pH reaches 7.0
- If the pH overgoes 7.0, add more solution B slowly until the pH backs to 7.0
- Solution A: dissolve 17.418 g of K2HPO4 (FW = 174.18) in 1,000 ml in sterilized deionized water in sterilized bottle
Acknowledgments
The authors are grateful for the financial supports by the National Natural Science Foundation of China (No. 21307095, No. 21407061), the International Science & Technology Cooperation Program of China (No. 2016YFE0123700), the Collaborative Innovation Center for Regional Environmental Quality, and the Swedish Research Council (contract Dnr. 639-2013-6913).
References
- Balaban, R. S., Nemoto, S. and Finkel, T. (2005). Mitochondria, oxidants, and aging. Cell 120(4): 483-495.
- Carretero, M., Solis, G.M., Petrascheck, M. (2017). C. elegans as model for drug discovery. Curr Top Med Chem 17: 1-10.
- Dengg, M. and van Meel, J. C. (2004). Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. J Pharmacol Toxicol Methods 50(3): 209-214.
- Feng, S., Cheng, H., Xu, Z., Shen, S., Yuan, M., Liu, J. and Ding, C. (2015). Thermal stress resistance and aging effects of Panax notoginseng polysaccharides on Caenorhabditis elegans. Int J Biol Macromol 81: 188-194.
- Solis, G. M. and Petrascheck, M. (2015). Measuring Caenorhabditis elegans life span in 96 well microtiter plates. J Vis Exp 49: e2496.
- Yu, Z., Chen, X., Zhang, J., Wang, R. and Yin, D. (2013a). Transgenerational effects of heavy metals on L3 larva of Caenorhabditis elegans with greater behavior and growth inhibitions in the progeny. Ecotoxicol Environ Saf 88: 178-184.
- Yu, Z., Sun, G., Liu, Y., Yin, D. and Zhang, J. (2017). Trans-generational influences of sulfamethoxazole on lifespan, reproduction and population growth of Caenorhabditis elegans. Ecotoxicol Environ Saf 135: 312-318.
- Yu, Z. Y., Zhang, J. and Yin, D. Q. (2012). Toxic and recovery effects of copper on Caenorhabditis elegans by various food-borne and water-borne pathways. Chemosphere 87(11): 1361-1367.
- Yu, Z., Zhang, J., Chen, X., Yin, D. and Deng, H. (2013b). Inhibitions on the behavior and growth of the nematode progeny after prenatal exposure to sulfonamides at micromolar concentrations. J Hazard Mater 250-251: 198-203.
- Yu, Z., Zhang, J. and Yin, D. (2016). Multigenerational effects of heavy metals on feeding, growth, initial reproduction and antioxidants in Caenorhabditis elegans. PLoS One 11(4): e0154529.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, J., Chen, R., Yu, Z. and Xue, L. (2017). Superoxide Dismutase (SOD) and Catalase (CAT) Activity Assay Protocols for Caenorhabditis elegans. Bio-protocol 7(16): e2505. DOI: 10.21769/BioProtoc.2505.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









