- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Structural Analysis of Target Protein by Substituted Cysteine Accessibility Method
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2470 Views: 6704
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Protocol to Purify Human Mediator Complex From Freestyle 293-F Cells
Hui-Chi Tang [...] Ti-Chun Chao
Feb 20, 2025 2149 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2277 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views
Abstract
Substituted Cysteine Accessibility Method (SCAM) is a biochemical approach to investigate the water accessibility or the spatial distance of particular cysteine residues substituted in the target protein. Protein topology and structure can be annotated by labeling with methanethiosulfonate reagents that specifically react with the cysteine residues facing the hydrophilic environment, even within the transmembrane domain. Cysteine crosslinking experiments provide us with information about the distance between two cysteine residues. The combination of these methods enables us to obtain information about the structural changes of the target protein. Here, we describe the detailed protocol for structural analysis using SCAM.
Keywords: CysteineBackground
Structural analyses provide the critical information about the function of a target protein. X-ray crystallography and nuclear magnetic resonance have been utilized as high-resolution protein structural analysis methods in the biology field. However, these methods require a purified protein extracted from membrane at very high concentration for the structural analysis of membrane proteins. Substituted Cysteine Accessibility Method (SCAM) is a biochemical approach to analyze the water accessibility and the spatial distance of particular cysteine residues substituted in the target protein. Using methanethiosulfonate (MTS) reagents that specifically react with the cysteine residues facing the hydrophilic environment, we can annotate the topology and structure of the target protein. As the labeling reagent N-biotinylaminoethyl methanethiosulfonate (MTSEA-biotin) is impermeable to the plasma membrane (Seal et al., 1998), in intact cells, only extracellular, but not intracellular, cysteine residues are biotinylated (Figure 1A). In contrast, both extracellular and intracellular cysteine residues are exposed to the hydrophilic environment in microsomes and are biotinylated by MTSEA-biotin (Figure 1B). Moreover, the accessibility of cysteine can be analyzed by competition experiments using membrane-impermeable MTS derivatives (Figure 2A). By combining the results of these analyses, we are able to obtain structural information of the entire intrinsic protein in the membrane (Akabas et al., 1992; Loo and Clarke, 1995; Frillingos et al., 1998; Karlin and Akabas, 1998; Foucaud et al., 2001; Kaback et al., 2001; Bogdanov et al., 2005; Sato et al., 2006 and 2008; Takagi et al., 2010; Watanabe et al., 2010; Takagi-Niidome et al., 2015; Tominaga et al., 2016; Cai et al., 2017; Tomita, 2017). The accessibility of cysteine can be analyzed in more detail by competition experiments using membrane-impermeable MTS derivatives; the negatively charged 2-sulfonatoethyl methanethiosulfonate (MTSES), the positively charged 2-(trimethylammonium)-ethyl methanethiosulfonate (MTSET), and the sterically bulkiest 2-(Triethylammonium)-Ethyl Methanethiosulfonate Bromide (MTS-TEAE) (Figure 2A). Moreover, cross-linking experiments using microsome fractions provide information regarding the spatial distance between two separated cysteines in different polypeptides (Figure 3A) (Loo and Clarke, 1996; 2000; 2001; Klco et al., 2003). In this experiment, all endogenous cysteines of the target protein should be mutated by serine or alanine (cys-less mutant), and then one (single cys-mutation) or two (double cys-mutation) target residues would be substituted to the cysteine to analyze the accessibility of particular residue(s) by MTS reagents. However, sometimes these substitutions affect the structure and function of the target protein. Thus, it is important to analyze the biological function of the target protein to ensure whether the mutations do not affect the protein conformation.
Materials and Reagents
- QSP 0.1-10 μl PIPETTE TIP (Thermo Fisher Scientific, Quality Scientific Plastics, catalog number: 102-Q )
- 1-200 μl New Pipette Tip Yellow (Thermo Fisher Scientific, Quality Scientific Plastics, catalog number: TW110-Q )
- 101-1,000 μl Pipette Tip Blue (Thermo Fisher Scientific, Quality Scientific Plastics, catalog number: 111-Q )
- 1,000-5,000 μl Pipette Tip Natural (Thermo Fisher Scientific, Quality Scientific Plastics, catalog number: 090-Q )
- 15 ml centrifuge tube (IWAKI, catalog number: 2325-015 )
- 12-well flat bottom microplate (IWAKI, catalog number: 3815-012 )
- 150 mm tissue culture dish (IWAKI, catalog number: 3030-150 )
- 1 ml syringe (TERUMO, catalog number: SS-01T )
- 27 G needle (TERUMO, catalog number: NN-2719S )
- MEF cells
- Immunostar® Reagents (Immunostar; Wako Pure Chemical Industries, catalog number: 291-55203 )
- SuperSignalTM West Femto Maximum Sensitivity Substrate (Supersignal; Thermo Fisher Scientific, catalog number: 34096 )
- Streptavidin SepharoseTM High Performance (SA beads; GE Healthcare, catalog number: 17511301 )
- Disodium hydrogenphosphate 12-Water (Na2HPO4•12H2O; Wako Pure Chemical Industries, catalog number: 196-02835 )
- Sodium dihydrogenphosphate Dihydrate (NaH2PO4•2H2O; Wako Pure Chemical Industries, catalog number: 192-02815 )
- Potassium chloride (KCl; Wako Pure Chemical Industries, catalog number: 163-03545 )
- Sodium chloride (NaCl; NACALAI TESQUE, catalog number: 31320-34 )
- Potassium dihydrogen phosphate (KH2PO4; Wako Pure Chemical Industries, catalog number: 169-04245 )
- Copper(II) sulfate (CuSO4; Wako Pure Chemical Industries, catalog number: 034-04445 )
- Sodium Dodecyl Sulfate (SDS; NACALAI TESQUE, catalog number: 31606-04 )
- 1,10-Phenanthroline monohydrate (Phenanthroline; Wako Pure Chemical Industries, catalog number: 169-00862 )
- N-Ethylmaleimide (NEM; NACALAI TESQUE, catalog number: 15512-11 )
- cOmpleteTM Protease inhibitor cocktail (Roche Diagnostics, catalog number: 11836145001 )
- 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES; Dojindo Molecular Technologies, catalog number: GB10 )
- O,O'-Bis(2-aminoethyl)ethyleneglycol-N,N,N',N'-tetraacetic acid (EGTA; Dojindo Molecular Technologies, catalog number: G002 )
- Sucrose (Wako Pure Chemical Industries, catalog number: 196-00015 )
- Tris (hydroxymethyl) aminomethane (Tris; C4H11NO3; STAR CHEMICAL, catalog number: RSP-THA500G )
- Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
- Brilliant Green (Wako Pure Chemical Industries, catalog number: 021-02352 )
- Coomassie Brilliant Blue G-250 (CBB G-250; NACALAI TESQUE, catalog number: 09409-42 )
- DMEM, high glucose, no glutamate, no methionine, no cysteine (DMEM (cys-); Thermo Fisher Scientific, catalog number: 21013-024 )
- Dimethyl sulfoxide (DMSO; Wako Pure Chemical Industries, catalog number: 045-28335 )
- 2-mercaptoethanol (Wako Pure Chemical Industries, catalog number: 139-07525 )
- N-Biotinylaminoethyl Methanethiosulfonate (MTSEA-biotin; Toronto Research Chemicals, catalog number: B394750 )
- Sodium 2-Sulfonatoethyl Methanethiosulfonate (MTSES; Toronto Research Chemicals, catalog number: S672000 )
- 2-(Trimethylammonium)-Ethyl Methanethiosulfonate Bromide (MTSET; Toronto Research Chemicals, catalog number: T795900 )
- 2-(Triethylammonium)-Ethyl Methanethiosulfonate Bromide (MTS-TEAE; Toronto Research Chemicals, catalog number: T775800 )
- 1,2-Ethanediyl Bismethanethiosulfonate (M2M; Toronto Research Chemicals, catalog number: E890350 )
- 1,3-Propanediyl Bismethanethiosulfonate (M3M; Toronto Research Chemicals, catalog number: P760350 )
- 1,4-Butanediyl Bismethanethiosulfonate (M4M; Toronto Research Chemicals, catalog number: B690150 )
- 1,6-Hexanediyl Bismethanethiosulfonate (M6M; Toronto Research Chemicals, catalog number: H294250 )
- 3,6-Dioxaoctane-1,8-diyl Bismethanethiosulfonate (M8M; Toronto Research Chemicals, catalog number: D486150 )
- Undecane-1,11-diyl-bismethanethiosulfonate (M11M; Toronto Research Chemicals, catalog number: U787800 )
- 3,6,9,12-Tetraoxatetradecane-1,14-diyl-bis-methanethiosulfonate (M14M; Toronto Research Chemicals, catalog number: T306250 )
- 3,6,9,12,15-Pentaoxaheptadecane-1,17-diyl Bis-methanethiosulfonate (M17M; Toronto Research Chemicals, catalog number: P273750 )
- 1x Phosphate Buffered Saline (1x PBS; see Recipes)
- 1x Dulbecco's Phosphate Buffered Saline (1x DPBS; see Recipes)
- 1x Sample buffer (sample buffer; see Recipes)
- 1x homogenize buffer (see Recipes)
Equipment
- 0.5-10 μl Pipettor (NICHIRYO, model: Nichipet EXII, catalog number: 00-NPX2-10 )
- 2-20 μl Pipettor (NICHIRYO, model: Nichipet EXII, catalog number: 00-NPX2-20 )
- 20-200 μl Pipettor (NICHIRYO, model: Nichipet EXII, catalog number: 00-NPX2-200 )
- 100-1,000 μl Pipettor (NICHIRYO, model: Nichipet EXII, catalog number: 00-NPX2-1000 )
- 1,000-5,000 μl Pipettor (NICHIRYO, model: Nichipet EXII, catalog number: 00-NPX2-5000 )
- Forma Series II Water Jacket CO2 Incubator (Thermo Fisher Scientific, catalog number: 3110 )
- Polytron homogenizer (Hitachi, model: HG30 )
- 500-Watt ultrasonic processor (Sonics & Materials, model: VCX 500 )
- Small size culture rotator (TAITEC, model: RT-50 )
- High speed centrifuge (Koki Holdings, himac, model: CF15RN )
- High speed refrigerated micro centrifuge (TOMY, model: MX-307 )
- Ultracentrifuge (Beckman Coulter, model: Optima L-90K )
- Ultracentrifuge fixed angle rotor (Beckman Coulter, model: Type 50.4 Ti , catalog number: 347299)
- Aluminum block bath (TAITEC, model: DTU-1CN )
- ImageQuant LAS 4000 (GE Healthcare)
- Centrifuge tube (Beckman Coulter, catalog number: 355645 )
- Spatula
Software
- ImageQuant TL 7.0 (GE Healthcare)
Procedure
- SCAM using intact MEF cells
- Seed 1 x 106 cells expressing target protein with single cys-mutation to 12-well plates, and culture for 12 h to reach 100% confluency.
- Wash the cells once with 1x PBS.
- For the labeling assay, skip Steps A3 and A4. For the competition assay using MTSES, MTSET, or MTS-TEAE, add competitors at a final concentration of 2 mM to each well. Incubate the cells on ice for 30 min under light-shielded conditions.
- Wash cells that are not being incubated with labeling compound once with 1x PBS. Make sure not to wash the cells that are being incubated with compound.
- Dilute 50 mM MTSEA-biotin with DMEM (cys-) at a final concentration of 1 mM. Add 450 μl DMEM (cys-) solution to each well and incubate the cells on ice for 30 min under light-shielded conditions.
- Wash the cells twice with 1x PBS. Take care not to remove the cells by the water pressure.
- Solubilize the cells with 500 μl/well of 1% SDS/PBS. Shake the plate for 30 min at room temperature.
- Sonicate the sample (pulse on/off time: 1 sec; amplitude: 35%) for 2 min 30 sec on ice. Make sure to sonicate the sample to the point where the sample is no longer stringy.
- Wash the SA beads 4 times using 1x DPBS. Elute the SA beads in 1x DPBS in the ratio of 66% SA beads to 34% 1x DPBS.
- Add 30 μl of the washed SA beads to each sample. Rotate overnight at room temperature.
- Centrifuge the SA beads at 12,900 x g for 45 sec at 4 °C. Remove the supernatant and add 500 μl of 1% SDS/PBS. Repeat 4 times.
- Elute samples by boiling in sample buffer containing 1% 2-mercaptoethanol at 100 °C for 1 min. Analyze the eluted proteins by immunoblotting. Sometimes the separation of the precipitated beads and the supernatant is difficult because the stickiness of the samples. In this case, refer to Note 3.
- Preparation of MEF microsomes
- Seed the cells expressing target protein with single/double Cys mutant onto a 15-cm dish and culture the cells until approximately 90% confluency.
- Wash the cells with an adequate amount of 1x PBS, and collect the cells in 5 ml/plate of 1x DPBS using a spatula. Cells from two plates are gathered into one 15-ml tube and pelleted by centrifugation at 750 x g for 10 min at 4 °C and stored at -80 °C until use.
- Suspend the pellet in 2 ml of 1x homogenization buffer containing cOmpleteTM Protease inhibitor cocktail. Cells are then transferred to centrifuge tubes and suspended using a 27 G needle. Homogenize the sample using a polytron homogenizer (speed 4) for 1 min on ice. Then ultracentrifuge the sample at 1,400 x g for 10 min at 4 °C. Add 2 ml of 1x homogenization buffer containing cOmpleteTM Protease inhibitor cocktail to the pellet. Homogenize the sample again at speed 4 for 1 min on ice. Ultracentrifuge the sample at 1,400 x g for 10 min at 4 °C. Combine the supernatant and ultracentrifuge at 29,000 x g for 40 min at 4 °C. Store the pellet at -80 °C until use.
- SCAM analysis using MEF microsomes
- Suspend microsomes expressing single cys-mutant in 200 μl of 1x DPBS with a 27 G needle to make 1 mg/ml protein solution.
- For the normal labeling assay, skip Step C2 and go on to Step C3. For the competition assay using MTSES, MTSET, or MTS-TEAE, add competitors at a final concentration of 2 mM to each well. Incubate the cells on ice for 30 min under light-shielded conditions.
- Dilute 50 mM MTSEA-biotin to the microsome solution at a final concentration of 1 mM. Incubate the microsomes on ice for 5 min under light-shielded conditions.
- Ultracentrifuge the sample at 12,900 x g for 5 min at 4 °C. Discard the supernatant and add 200 μl of 1x DPBS to the pellet and ultracentrifuge under the same conditions.
- Add 500 μl of 1% SDS/PBS to the pellet. Resuspend the pellet by gentle pipetting and solubilize by rotation at room temperature for 30 min.
- After the rotation, precipitate the biotinylated proteins using SA beads in the same manner to that of SCAM using intact cells (see Procedure A, Steps 9-11). Analyze the eluted proteins by immunoblotting.
- Crosslinking experiments using MTS crosslinkers
- Suspend microsomes expressing double cys-mutant in 1x DPBS and further suspend with a 27 G needle. The amount of 1x DBPS can be adjusted according to the expression level of the Cys mutant protein.
- Add 8 mM MTS crosslinker dissolved in DMSO into the microsome solution at a final concentration of 0.2 mM. Incubate the sample in an aluminum block bath at 37 °C for 30 min.
- Add 5 M NEM dissolved in DMSO to the sample at a final concentration of 10 mM to modify cysteine residues that did not react with the crosslinker.
- Add sample buffer without 2-mercaptoethanol, gently mix the sample using a vortex, and analyze by immunoblotting.
- Cross-linking experiments using copper-phenanthroline
- Microsomes are prepared as Step D1.
- Add 120 mM CuSO4 and 500 mM phenanthroline dissolved in DMSO to the microsome solution at final concentrations of 3 mM and 15 mM, respectively. Incubate the samples at room temperature for 2 h.
- Samples are analyzed as in Steps D3 and D4.
Data analysis
- The intensity of the bands obtained from immunoblotting is visualized by ImageQuant LAS 4000 and analyzed by ImageQuant TL. The loading volumes of the eluted proteins must be normalized by total protein concentration. An example of the bands obtained in the SCAM experiments is shown in Figure 1C.
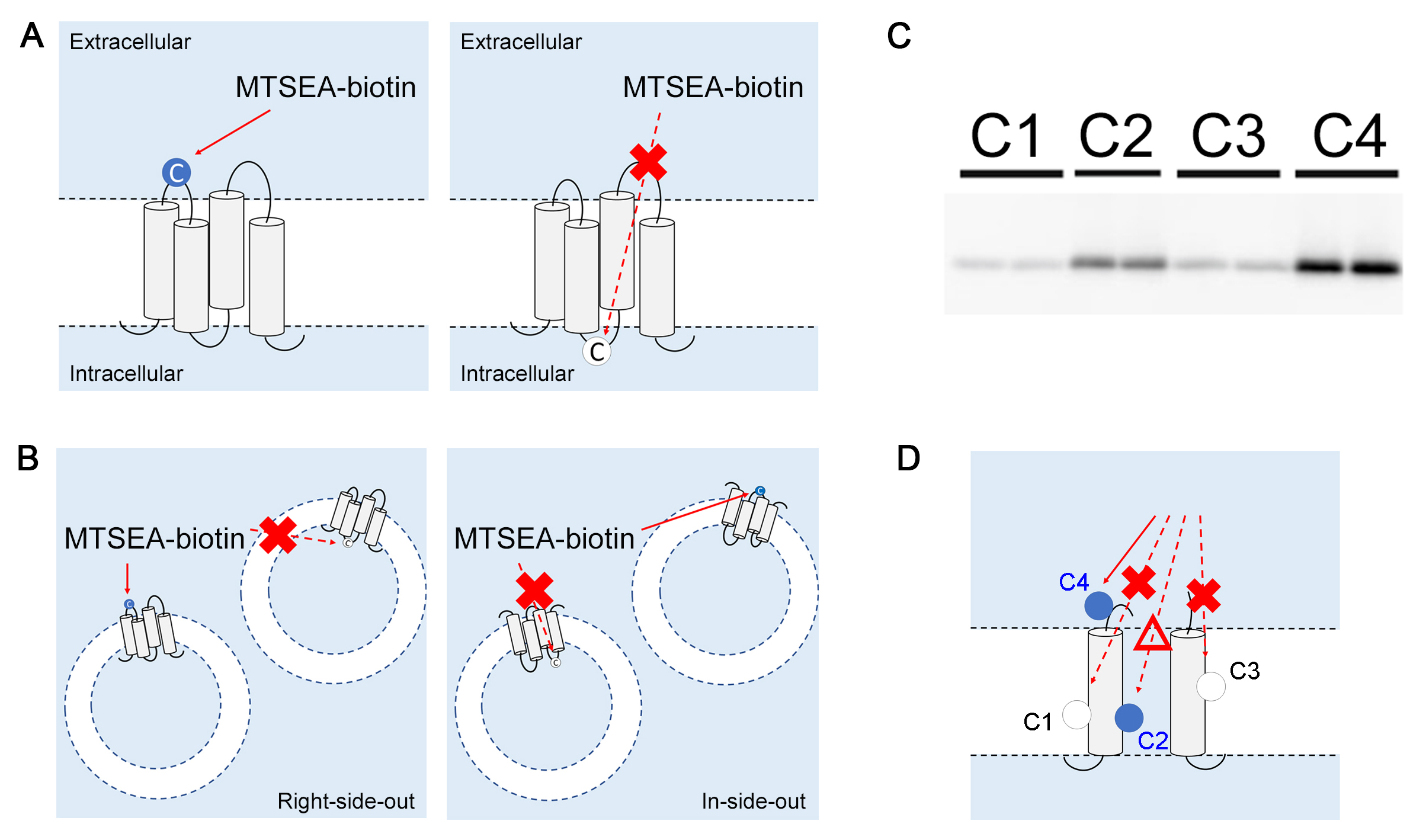
Figure 1. SCAM for topology mapping. A. In intact cells, only cysteine residues facing the extracellular region are labeled. B. In microsome fractions, cysteine residues facing both extracellular and intracellular sides are labeled. C. Duplicated results of SCAM for cysteines facing different hydrophilic environments. The labeling efficiency by MTSEA-biotin reflects the water accessibility of the target residue. Thus, higher intensity of the band represents the higher the water accessibility (i.e., C4 > C2 > C1 = C3). D. Representative image of the 4 cysteines in C. Dash lines indicate less accessibility of the MTS reagent to the designated residues (i.e., C1, C2 and C3) compared to that to C4. MTS reagent has the weakest accessibility to C1 and C3 (cross symbols) whereas accessibility to C2 is stronger though still weaker than C4 (triangle symbol). - The MTS-based competitors MTSES, MTSET, and MTS-TEAE are different in size, in the order of (from largest to smallest) MTS-TEAE > MTSET > MTSES > MTSEA-biotin (Tominaga et al., 2016). The sizes and the ionic charges affect permeability into a narrow hydrophilic environment (Figure 2A). Thus, the competition of labeling between two MTS reagents reflects the condition of the hydrophilic environment around the target cysteine (Figure 2B).
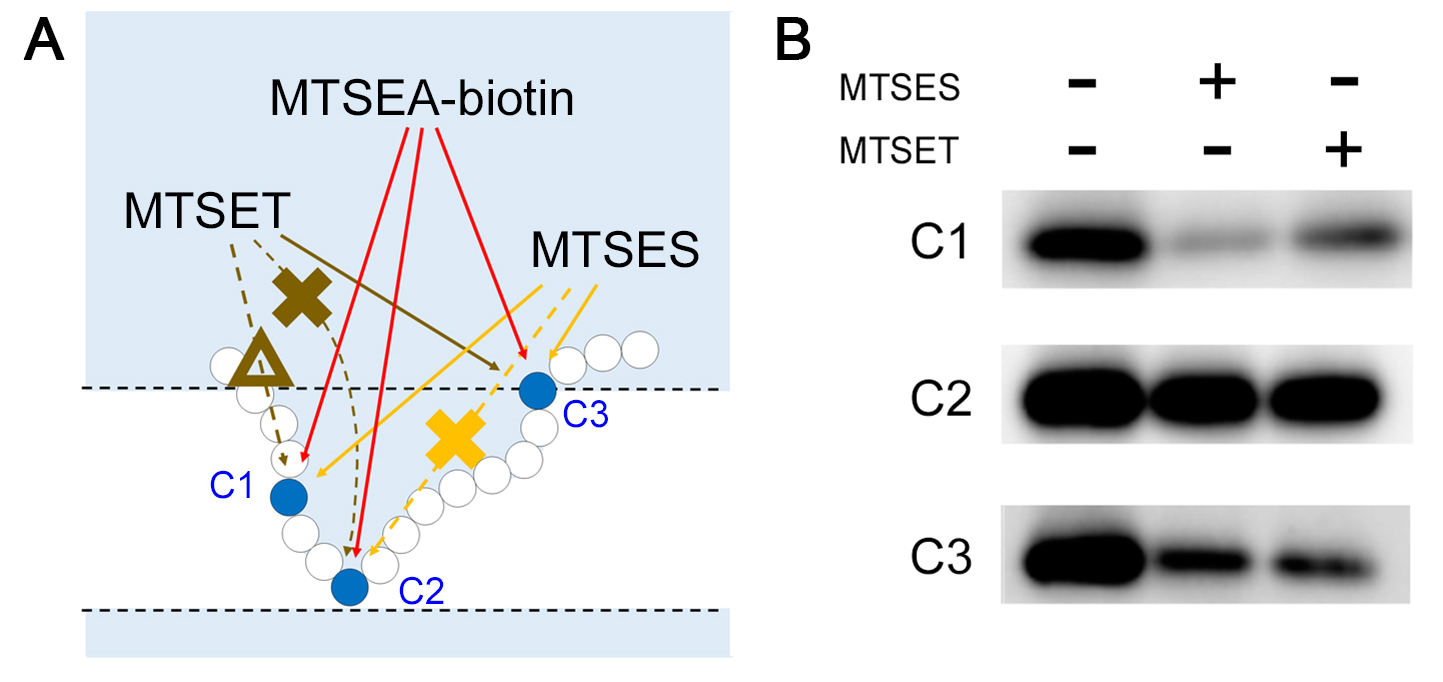
Figure 2. Competition assay using MTS reagents. A. Competition efficiency reflects the water accessibility of the cysteine. Dash lines indicate the reagent has less or no accessibility to the cysteine. If cysteine residues are facing an open hydrophilic environment (C1 and C3), labeling by MTSEA-biotin is decreased by preincubation of either of the MTS reagents (i.e., MTSET or MTSES). However, MTSET is less accessible to C1 than MTSES (triangle symbol), different competition efficiency is observed. For cysteines located in the restricted hydrophilic environment (C2), both MTS reagents fail to react with the target cysteine (cross symbol). B. An example of the results of labeling and competition of cysteines facing different hydrophilic environments (C1-C3 in [A]). - Crosslinking experiments with compounds carrying different spacer arms are used for measuring the spatial distance between two separated proteins (Figure 3A). Crosslinked proteins are detected by the appearance of a band corresponding to the protein heterodimer, accompanied by a reduction in the amount of each monomer protein (Figure 3B). Eight MTS-derived cross-linkers (M2M, M3M, M4M, M6M, M8M, M11M, M14M, and M17M, with spacer arms of 5.2, 6.5, 7.8, 10.4, 13.0, 16.9, 20.8, and 24.7 angstroms, respectively) are used in the crosslinking experiments using microsome fractions (Loo and Clarke, 2001). Copper and phenanthroline, which are redox catalysts that oxidize free sulfhydryls and form disulfide bonds in cysteines that collide can be applied in this method (Klco et al., 2003).
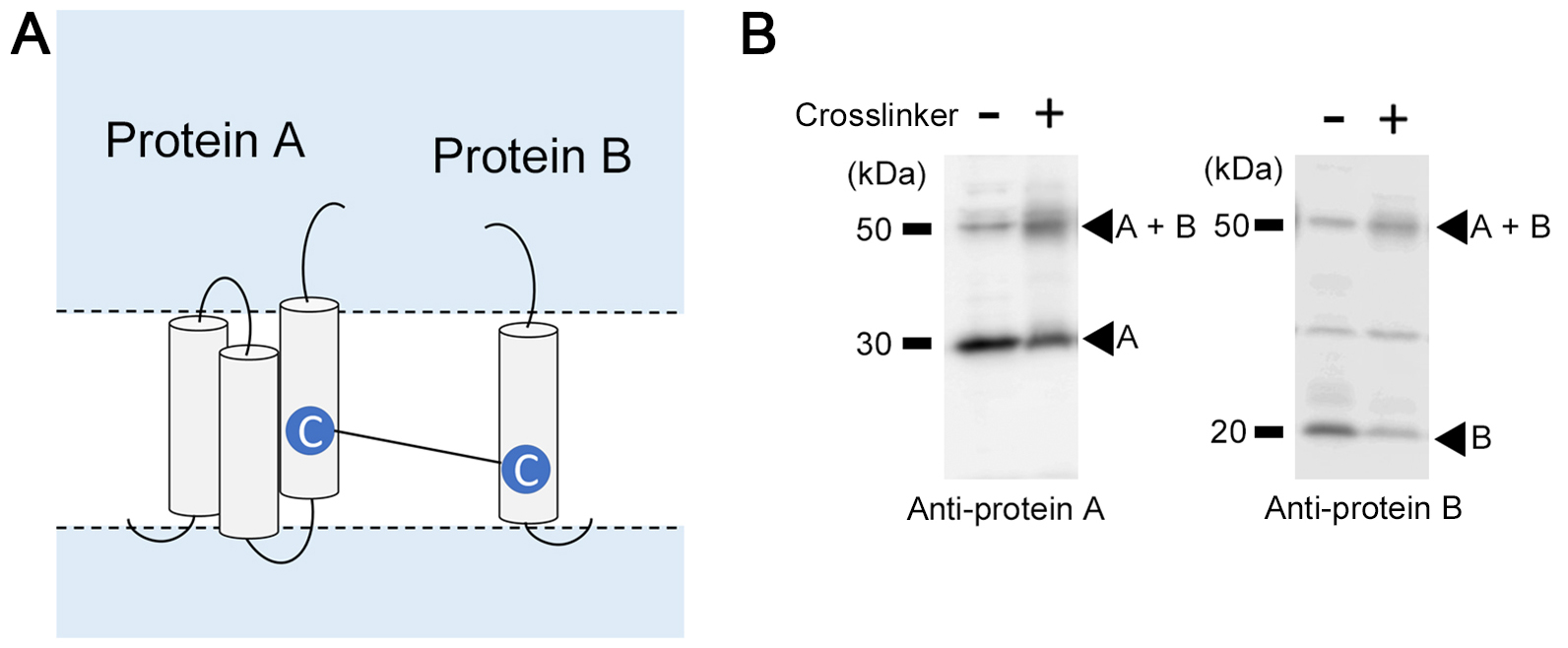
Figure 3. Crosslinking experiments. A. Two cysteines in different polypeptides are crosslinked with MTS-based crosslinkers or copper and phenanthroline. B. Results of crosslinking experiments between proteins A and B.
Notes
- It is very important to check the expression level of the target protein by appropriate methods before SCAM.
- MTSEA-biotin should be dissolved in DMSO and dispensed before use in each experiment to avoid repeated freeze-thaw cycles.
- If SA beads stick to the samples, the following additional procedures can be tested: 1) prepare 1% SDS/PBS for solubilization of the cells and wash the beads before each use, 2) sonicate the samples longer to reduce the viscosity of the sample, 3) homogenize the sample by vortexing during the sonication step, and 4) reduce the time of rotation of the sample (3 h minimum).
- Reducing agents, such as 2-mercaptoethanol or boiling the samples at high temperatures are strictly prohibited in the analysis of crosslinked proteins.
- The protein solution at the concentration of 1 mg/ml is one example. The protein concentration can be increased or reduced according to the expression level of the target protein.
- This protocol is designed for MEF cells. However, other cell types (e.g., HEK 293 cells) have been applied to SCAM to analyze various membrane proteins such as dopamine receptor and acetylcholine receptor. In such cases, experimental conditions including protein concentration, compound incubation time must be optimized.
- Crosslinked proteins should be analyzed by immunoblotting on the day of the experiment.
- For quantitative analysis of the bands, chemiluminescence reagents and exposure times are crucial to avoid saturation of band intensities. We recommend the use of reagents with weaker reactivity (e.g., Immunostar) first, then test reagents with higher reactivity (e.g., Supersignal).
Recipes
- 1x PBS
8 mM Na2HPO4•12H2O
2 mM NaH2PO4•2H2O
131 mM NaCl in distilled water - 1x DPBS
6 mM NaCl
0.32 mM Na2HPO4•12H2O
108 μM KCl
60 μM KH2PO4 in distilled water - 1x sample buffer
2% SDS
0.08 M Tris-HCl (pH 6.8)
10% Glycerol
0.2% Brilliant Green
0.2% CBB-G250 in distilled water - 1x homogenize buffer
20 mM HEPES-NaOH pH 7.0
140 mM KCl
0.25 M Sucrose
1 mM EGTA in distilled water
Acknowledgments
We are grateful to Drs. Bart De Strooper (KU Leuven, Belgium) and Toshio Kitamura (The University of Tokyo, Japan) for valuable reagents used in SCAM. We would like to thank Dr. Takeshi Iwatsubo for longtime support with our SCAM study. This work was supported in part by a Grant-in-Aid for Scientific Research (A) (grant No. 15H02492 to TT) and Research Fellowships for young scientists (grant No. 18J14653 to TC) from the Japan Society for the Promotion of Science (JSPS). Part of figures are adapted and modified from the study of Cai et al. (2017).
Competing interests
The authors declare there are no potential conflicts of interest.
References
- Akabas, M. H., Stauffer, D. A., Xu, M. and Karlin, A. (1992). Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science 258(5080): 307-310.
- Bogdanov, M., Zhang, W., Xie, J. and Dowhan, W. (2005). Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAMTM): application to lipid-specific membrane protein topogenesis. Methods 36(2): 148-171.
- Cai, T., Yonaga, M. and Tomita, T. (2017). Activation of γ-secretase trimming activity by topological changes of transmembrane domain 1 of presenilin 1. J Neurosci 37(50): 12272-12280.
- Foucaud, B., Perret, P., Grutter, T. and Goeldner, M. (2001). Cysteine mutants as chemical sensors for ligand-receptor interactions. Trends Pharmacol Sci 22(4): 170-173.
- Frillingos, S., Sahin-Toth, M., Wu, J. and Kaback, H. R. (1998). Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J 12(13): 1281-1299.
- Kaback, H. R., Sahin-Toth, M. and Weinglass, A. B. (2001). The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol 2(8): 610-620.
- Karlin, A. and Akabas, M. H. (1998). Substituted-cysteine accessibility method. Methods Enzymol 293: 123-145.
- Klco, J. M., Lassere, T. B. and Baranski, T. J. (2003). C5a receptor oligomerization. I. Disulfide trapping reveals oligomers and potential contact surfaces in a G protein-coupled receptor. J Biol Chem 278(37): 35345-35353.
- Loo, T. W. and Clarke, D. M. (1995). Membrane topology of a cysteine-less mutant of human P-glycoprotein. J Biol Chem 270(2): 843-848.
- Loo, T. W. and Clarke, D. M. (1996). Inhibition of oxidative cross-linking between engineered cysteine residues at positions 332 in predicted transmembrane segments (TM) 6 and 975 in predicted TM12 of human P-glycoprotein by drug substrates. J Biol Chem 271(44): 27482-27487.
- Loo, T. W. and Clarke, D. M. (2000). Drug-stimulated ATPase activity of human P-glycoprotein is blocked by disulfide cross-linking between the nucleotide-binding sites. J Biol Chem 275(26): 19435-19438.
- Loo, T. W. and Clarke, D. M. (2001). Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. J Biol Chem 1276(40): 36877-36880.
- Sato, C., Morohashi, Y., Tomita, T. and Iwatsubo, T. (2006). Structure of the catalytic pore of γ-secretase probed by the accessibility of substituted cysteines. J Neurosci 26(46): 12081-12088.
- Sato, C., Takagi, S., Tomita, T. and Iwatsubo, T. (2008). The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the γ-secretase. J Neurosci 28(24): 6264-6271.
- Seal, R. P., Leighton, B. H. and Amara, S. G. (1998). Transmembrane topology mapping using biotin-containing sulfhydryl reagents. Methods Enzymol 296: 318-331.
- Takagi-Niidome, S., Sasaki, T., Osawa, S., Sato, T., Morishima, K., Cai, T., Iwatsubo, T. and Tomita, T. (2015). Cooperative roles of hydrophilic loop 1 and the C-terminus of presenilin 1 in the substrate-gating mechanism of γ-secretase. J Neurosci 35(6): 2646-2656.
- Takagi, S., Tominaga, A., Sato, C., Tomita, T. and Iwatsubo, T. (2010). Participation of transmembrane domain 1 of presenilin 1 in the catalytic pore structure of the γ-secretase. J Neurosci 30(47): 15943-15950.
- Tominaga, A., Cai, T., Takagi-Niidome, S., Iwatsubo, T. and Tomita, T. (2016). Conformational changes in transmembrane domain 4 of presenilin 1 are associated with altered amyloid-β 42 production. J Neurosci 36(4): 1362-1372.
- Tomita, T. (2017). Probing the structure and function relationships of presenilin by substituted-cysteine accessibility method. Methods Enzymol 584: 185-205.
- Watanabe, N., Takagi, S., Tominaga, A., Tomita, T. and Iwatsubo, T. (2010). Functional analysis of the transmembrane domains of presenilin 1: participation of transmembrane domains 2 and 6 in the formation of initial substrate-binding site of γ-secretase. J Biol Chem 285(26): 19738-19746.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Cai, T. and Tomita, T. (2018). Structural Analysis of Target Protein by Substituted Cysteine Accessibility Method. Bio-protocol 8(17): e2470. DOI: 10.21769/BioProtoc.2470.
- Cai, T., Yonaga, M. and Tomita, T. (2017). Activation of γ-secretase trimming activity by topological changes of transmembrane domain 1 of presenilin 1. J Neurosci 37(50): 12272-12280.
Category
Biochemistry > Protein > Structure
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








