- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Membrane Lipid Screen to Identify Molecular Targets of Biomolecules
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2427 Views: 9712
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2441 Views

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1600 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 482 Views
Abstract
Proteins that bind to and disrupt cell membranes may target specific phospholipids. Here we describe a protocol to identify the lipid targets of proteins and biomolecules. First, we describe a screen to identify lipids in membranes that are specifically bound by the biomolecule of interest. Second, we describe a method for determining if the presence of these lipids within membranes is necessary for membrane disruption. The methods described here were used to determine that the malaria vaccine candidate CelTOS disrupts cell membranes by specifically targeting phosphatidic acid (Jimah et al., 2016). This protocol has a companion protocol: ‘Liposome disruption assay to examine lytic properties of biomolecules’ which can be applied to examine the ability of the biomolecule to disrupt membranes composed of the lipid target identified by following this protocol (Jimah et al., 2017).
Keywords: MembraneBackground
Proteins and biomolecules with membrane disruption activities, such as pore formation or membrane fusion, may target specific lipids within membranes. Examples of lipid-specific pore-formation include Plasmodium CelTOS that depends on phosphatidic acid for pore formation, and the cholesterol dependent cytolysins (Jimah et al., 2016; Lukoyanova et al., 2016). CelTOS (cell traversal protein for ookinetes and sporozoites) is a malaria parasite protein that disrupts host cell membranes by pore formation to enable the exit of parasites from invaded host cells during cell traversal (Kariu et al., 2006; Jimah et al., 2016). Cholesterol dependent cytolysins are a large class of pore-forming proteins, including virulence factors of gram positive bacteria such as pneumolysin and listeriolysin (Lukoyanova et al., 2016). Identifying the specific lipids targeted informs the mechanism of membrane disruption that underlies biological function and role of proteins and biomolecules.
Materials and Reagents
- Small gel incubation tray (Santa Cruz Biotechnology, catalog number: sc-358889 )
- Serological pipets, 10 ml (Genesee Scientific, catalog number: 12-104 )
- Pipette tips
10 µl tips (VWR, catalog number: 46620-318 )
200 µl tips (VWR, catalog number: 53509-009 )
1,000 µl tips (VWR, catalog number: 83007-384 ) - Centrifuge tubes, 50 ml (Genesee Scientific, catalog number: 21-106 )
- Microcentrifuge tubes, 1.7 ml (MIDSCI, catalog number: AVSS1700RA )
- Membrane lipid strips with dimensions 2 x 3 cm (Echelon Biosciences, catalog number: P-6002 )
- Protein or biomolecule of interest
- Tris (Gold Bio, catalog number: T-400-5 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888-25KG )
- Tween 20% (Sigma-Aldrich, catalog number: P1379 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906-500G )
- Primary antibody against the tagged protein or biomolecule of interest
- Peroxidase conjugated secondary antibody
- ECL Prime Western Blotting Detection Kit (GE Healthcare, catalog number: RPN2232 )
- Phospholipids (dissolved in chloroform, commonly used phospholipids are):
DOPC (Avanti Polar Lipids, catalog number: 850375C )
POPC (Avanti Polar Lipids, catalog number: 850457C )
POPA (Avanti Polar Lipids, catalog number: 840857C )
POPS (Avanti Polar Lipids, catalog number: 840034C ) - Blocking buffer (see Recipes)
- Wash buffer (see Recipes)
Note: See the ‘Notes’ section for a list of materials and reagents used in the companion protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’ that is recommended for follow up experiments (Jimah et al., 2017).
Equipment
- Pipetman Classic pipets
P10 (Gilson, catalog number: F144802 )
P20 (Gilson, catalog number: F123600 )
P200 (Gilson, catalog number: F123601 )
P1000 (Gilson, catalog number: F123602 ) - Pipet-Aid XP Pipette controller (Drummond Scientific, catalog number: 4-000-101 )
- Incubator
- BenchRockerTM variable 2D rocker (Benchmark Scientific, catalog number: BR2000 )
- pH meter (Fisher Scientific, model: AccumetTM AE150, catalog number: 13-636-AE153 )
- Fluorescent image analyzer (Fujifilm, model: FLA-5000 )
Note: See the ‘Notes’ section for a list of equipment used in the companion protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’ that is recommended for follow up experiments (Jimah et al., 2017).
Procedure
Screen to identify specific membrane lipid binding by biomolecules of interest
- Purify or obtain the biomolecule of interest.
Note: Ensure the protein is tagged, for example with a 6-His tag. - Place lipid strips in a small gel incubation tray, and block lipid strips with 10 ml blocking buffer (see Recipes) at room temperature for one hour.
Notes: - See the ‘Notes’ section for a description of the lipid component of the lipid strips.
- The buffer and pH used depend on the biomolecule tested. For example, acidic conditions could be used if the biomolecule functions in an acidic environment. Also, use enough buffer to cover the lipid strip.
- Wash three times, for five minutes each, with 10 ml wash buffer (see Recipes).
Note: Suggestions on the duration of washing, blocking and incubation times is described in the Notes section. - Incubate the membrane lipid strips with the protein or biomolecule in 10 ml blocking buffer for one hour at room temperature.
Note: The concentration of protein used is determined empirically, and is usually at the low micromolar to nanomolar concentrations. - Wash three times, for five minutes each, with 10 ml wash buffer.
- Incubate the membrane lipid strips with a primary antibody against the tagged protein or biomolecule in 10 ml blocking buffer for one hour at room temperature.
Notes: - The primary antibody could be against the protein tag or specific to the protein or biomolecule.
- The concentration of primary antibody used is specified by the manufacturer.
- Wash three times, for five minutes each, with 10 ml wash buffer.
- Incubate the membrane lipid strips with a peroxidase conjugated secondary antibody, against the primary antibody, in 10 ml blocking buffer for one hour at room temperature.
Note: The concentration of secondary antibody used is specified by the manufacturer. - Use the ECL Prime Western Blotting Detection Kit to detect the peroxidase conjugated secondary antibody.
- Image the chemiluminescence using a fluorescent image analyser (Figure 1).
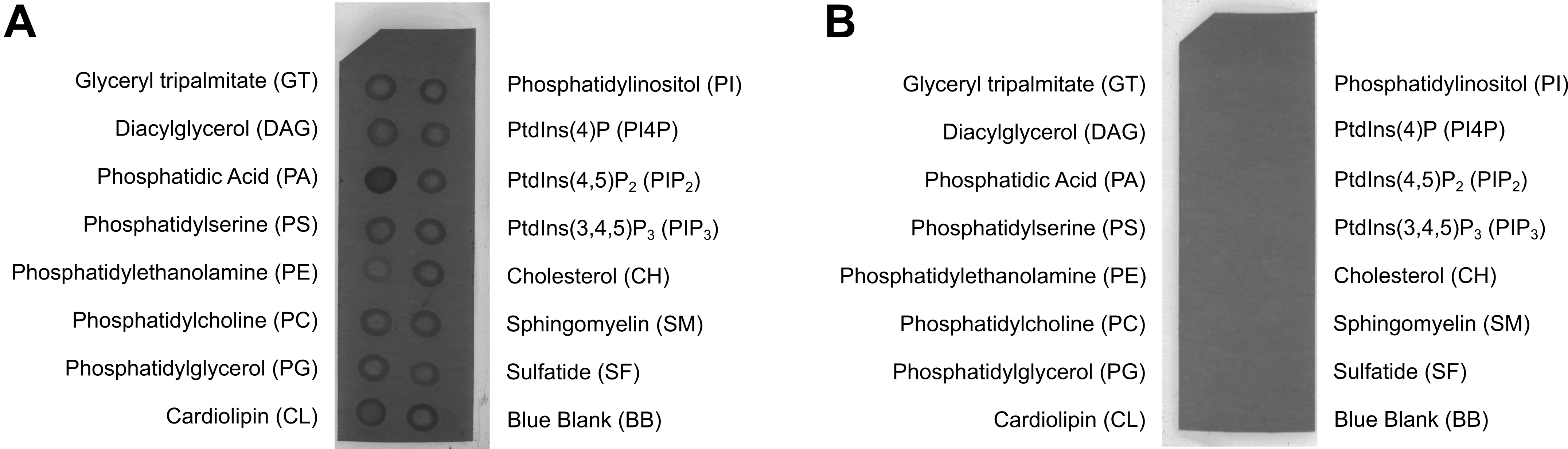
Figure 1. Lipid strips probed with PfCelTOS or controls. A. Lipid strip probed with PfCelTOS, followed by primary antibody to CelTOS and a secondary antibody conjugated with peroxidase for visualization. PfCelTOS specifically bound to phosphatidic acid (Jimah et al., 2016). B. Lipid strip probed with primary antibody against His-tag and a secondary antibody conjugated with peroxidase for visualization. Neither the primary or secondary antibody bound to the lipid strip.
Data analysis
- Screen to identify specific membrane lipid binding by biomolecules of interest:
- Spot intensities in each strip are normalized to the background for that strip and to the intensities in the negative control(s).
Note: The negative control may be a no protein control, or a protein that does not bind lipids. - The normalized spot intensities, from experimental replicates, are compared to determine if there is a statistical preference for specific lipids by one-way ANOVA.
Note: Perform appropriate number of replicates for statistical analysis. An example of representative data and analysis is reported in Jimah et al., 2016.
- Spot intensities in each strip are normalized to the background for that strip and to the intensities in the negative control(s).
- Conclusion
The protocol described here enables the identification of specific lipids that are targeted by proteins of biomolecules for binding. In addition to binding lipid molecules, some proteins or biomolecules may disrupt membranes by targeting a specific lipid component of a membrane. A companion protocol: ‘Liposome disruption assay to test lytic properties of biomolecules’ describes how to determine and quantify the disruption of membranes by proteins or biomolecules (Jimah et al., 2017). Below is a summary of ‘Screen to identify membrane lipids targeted for membrane disruption by biomolecules’.
Procedure: Screen to identify membrane lipids targeted for membrane disruption by biomolecules
Notes:- The identification of specific lipids targeted for binding by biomolecules, described above, will inform the lipids targeted for membrane disruption since binding precedes membrane disruption.
- The protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’ will be applied to determine if the presence of a particular lipid within membranes is necessary for membrane disruption.
- Make liposomes composed of lipids that have been identified to bind the biomolecule of interest.
Note: For example, if the biomolecule binds phosphatidic acid, liposomes composed of phosphatidylcholine and phosphatidic acid at 8:2 molar ratio may be made. The addition of phosphatidylcholine is because liposomes composed of phosphatidic acid alone are unstable. In the case of this example, it is also useful to make liposomes composed only of phosphatidylcholine to serve as a negative control. - Investigate the membrane disruption activity of the biomolecule on the liposomes following the protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’.
- Test a range of concentrations of the biomolecule for the ability to disrupt liposomes.
Note: Recommended concentrations are within the nanomolar and micromolar range. The biomolecule targets specific lipids if low nanomolar concentrations of the biomolecule disrupt membranes containing particular lipids. It may be possible that at high concentrations, the biomolecule nonspecifically disrupts membranes composed of other lipids.Data analysis: Screen to identify membrane lipids targeted for membrane disruption by biomolecules
Note: Please refer to the companion protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’ for a detailed description of data analysis. (Jimah et al., 2017).
- The identification of specific lipids targeted for binding by biomolecules, described above, will inform the lipids targeted for membrane disruption since binding precedes membrane disruption.
Notes
- Membrane lipid strips (Echelon Biosciences) contain the common membrane lipids: triglyceride, phosphatidylinositol, phosphatidylinositol (4)-phosphate, phosphatidylinositol (4,5)-bisphosphate, phosphatidylinositol (3,4,5)-trisphosphate, phosphatidylserine, phosphatidylethanolamine, phosphatidic acid, diacylglycerol, cholesterol, phosphatidylcholine, sphingomyelin, phosphatidylglycerol, 3-sulfogalactosylceramide and cardiolipin.
- Another membrane lipid strip contains additional lipids that may be of interest (Echelon Biosciences, catalog number: P-6001) contains: Lysophosphatidic acid (LPA), Lysophosphocholine (LPC), Phosphatidylinositol (PtdIns), Phosphatidylinositol (3) phosphate (PtdIns(3)P), Phosphatidylinositol (4) phosphate (PtdIns(4)P), Phosphatidylinositol (5) phosphate (PtdIns(5)P), Phosphatidylethanolamine (PE), Phosphatidylcholine (PC), Sphingosine 1-Phosphate (S1P), Phosphatidylinositol (3,4) bisphosphate (PtdIns(3,4)P2), Phosphatidylinositol (3,5) bisphosphate (PtdIns(3,5)P2), Phosphatidylinositol (4,5) bisphosphate (PtdIns(4,5)P2), Phosphatidylinositol (3,4,5) trisphosphate (PtdIns(3,4,5)P3), Phosphatidic acid (PA), Phosphatidylserine (PS).
- The blocking, incubation, and washing times stated here are recommendations, and may be modified based on empirical observations.
- Please refer to the companion protocol ‘Liposome disruption assay to examine lytic properties of biomolecules’ for a detailed description of the materials, reagents, equipment and protocol necessary for the identification of membrane lipids targeted for membrane disruption by biomolecules (Jimah et al., 2017).
Recipes
- Blocking buffer
10 mM Tris pH 8.0
150 mM NaCl
0.1% Tween 20%
3% BSA - Wash buffer
10 mM Tris pH 8.0
150 mM NaCl
0.1% Tween 20%
Acknowledgments
This work was supported by the Burroughs Wellcome Fund (to NHT) and National Institutes of Health (R56 AI080792 to NHT). This protocol was adapted from Jimah et al., 2016.
References
- Jimah, J. R., Salinas, N. D., Sala-Rabanal, M., Jones, N. G., Sibley, L. D., Nichols, C. G., Schlesinger, P. H. and Tolia, N. H. (2016). Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption. Elife 5.
- Jimah, R. J., Schlesinger, H. P. and Tolia, H. N. (2017). Liposome disruption assay to examine lytic properties of biomolecules. Bio Protoc 7(15): e2433.
- Kariu, T., Ishino, T., Yano, K., Chinzei, Y. and Yuda, M. (2006). CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol 59(5): 1369-1379.
- Lukoyanova, N., Hoogenboom, B. W. and Saibil, H. R. (2016). The membrane attack complex, perforin and cholesterol-dependent cytolysin superfamily of pore-forming proteins. J Cell Sci 129(11): 2125-2133.
Article Information
Copyright
Jimah et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Jimah, J. R., Schlesinger, P. H. and Tolia, N. H. (2017). Membrane Lipid Screen to Identify Molecular Targets of Biomolecules. Bio-protocol 7(15): e2427. DOI: 10.21769/BioProtoc.2427.
- Jimah, J. R., Salinas, N. D., Sala-Rabanal, M., Jones, N. G., Sibley, L. D., Nichols, C. G., Schlesinger, P. H. and Tolia, N. H. (2016). Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption. Elife 5.
Category
Biochemistry > Protein > Activity
Microbiology > Microbe-host interactions > In vitro model
Biochemistry > Lipid > Lipid-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









