- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Polyamine and Paraquat Transport Assays in Arabidopsis Seedling and Callus
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2421 Views: 7809
Reviewed by: Marisa RosaJoëlle SchlapferAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Tensile Testing Assay for the Measurement of Tissue Stiffness in Arabidopsis Inflorescence Stem
Kouki Yoshida [...] Nobutaka Mitsuda
Aug 5, 2019 10162 Views

Analysis of Monosaccharides from Arabidopsis Seed Mucilage and Whole Seeds Using HPAEC-PAD
Gillian H. Dean [...] George W. Haughn
Dec 20, 2019 5744 Views

Targeting Ultrastructural Events at the Graft Interface of Arabidopsis thaliana by A Correlative Light Electron Microscopy Approach
Clément Chambaud [...] Lysiane Brocard
Jan 20, 2023 2973 Views
Abstract
Polyamines (PAs) are polycationic compounds found in all living organisms and play crucial roles in growth and survival. We here show the ‘Polyamine and paraquat (PQ) transport assay’ protocol, which can be used to examine the uptake activity of PA/PQ transporters. We have used this protocol to demonstrate that PUT3 in Arabidopsis is a polyamine transporter and is able to take up spermidine and its analog paraquat.
Keywords: Arabidopsis thalianaBackground
PAs are involved in gene regulation by interacting with and modulating the functions of anionic macromolecules such as DNA, RNA and proteins. In living cells, PAs’ contents must be regulated to maintain the cellular hemostasis. In higher plants, three major polyamines, putrescine (Put), spermidine (Spd) and spermine (Spm), are present in either free form or conjugated forms with other molecules (Gill and Tuteja, 2010). In yeast, four plasma membrane polyamine transporters, DUR3, SAM3, GAP1 and AGP2 were identified (Uemura et al., 2007). In Arabidopsis, five putative polyamine uptake transporters (PUT1-PUT5) were identified and PUT1-3 have been experimentally validated as polyamine transporters (Mulangi et al., 2012; Li et al., 2013). Our protocol described below has successfully confirmed that PUT3 is an influx transporter for polyamines and paraquat, and PQ/Spd uptake is impaired in the put3 mutant (Shen et al., 2016).
Materials and Reagents
- Pipette tips
- Plastic Petri dish (VWR, catalog number: 25384-326 )
- Parafilm
- 1.5 ml Eppendorf centrifuge tube (VWR, catalog number: 20170-355 )
- Filter paper
- Blue pestle (DWK Life Sciences, Kimble, catalog number: 749521-1500 )
- Cuvette (VWR, catalog number: 414004-051 )
- Syringe filters, 0.2 µm pore size (VWR, catalog number: 28145-475 )
- Syringe
- Arabidopsis thaliana ecotype Columbia (Col-0) and mutant line lhr1 (put3)
- ScintiVerseTM BD Cocktail (Fisher Scientific, catalog number: SX18-4 )
- Clorox Bleach
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- (2,4-Dichlorophenoxy) acetic acid sodium salt monohydrate (Sigma-Aldrich, catalog number: D6679 )
- Kinetin (Duchefa Biochemie, catalog number: K0905 )
- Sodium hydroxide (NaOH)
- Murashige & Skoog Basal Salt Mixture (PhytoTechnology Laboratories, catalog number: M524 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Low melting point agarose (Sigma-Aldrich, catalog number: A9414 )
- Methyl viologen dichloride hydrate (Sigma-Aldrich, catalog number: 856177 )
- Spermidine (MP Biomedicals, catalog number: 02152068 )
- Paraquat-methyl-14C dichloride hydrate (Sigma-Aldrich, catalog number: 313947 )
Note: This product has been discontinued. - Spermidine trihydrochloride [Terminal Methylenes-3H (N)] (PerkinElmer, catalog number: NET522001MC )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) (Sigma-Aldrich, catalog number: E5134 )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M7522 )
Note: This product has been discontinued. - Seed sterilization solution (see Recipes)
- 2,000x 2,4-D (9.05 mM) (see Recipes)
- 2,000x kinetin (1.86 mM) (see Recipes)
- Callus induction solid medium (see Recipes)
- Callus induction liquid medium (see Recipes)
- ½ MS solid medium (see Recipes)
- ½ MS liquid medium (see Recipes)
- 200 µM non-14C-labeled PQ solution (see Recipes)
- 2.04 mM non-3H-labeled Spd solution (see Recipes)
- 40.37 µM non-3H-labeled Spd solution (see Recipes)
- Solution 1: 14C-labeled PQ solution (see Recipes)
- Solution 2: 14C-labeled PQ solution (see Recipes)
- Solution 3: 3H-labeled Spd solution (see Recipes)
- Solution 4: 3H-labeled Spd solution (see Recipes)
- Solution 5: 3H-labeled Spd solution (see Recipes)
- Solution 6: 3H-labeled Spd solution (see Recipes)
- 1 M Tris-HCl, pH 7.5 (see Recipes)
- 0.5 M EDTA, pH 8.0 (see Recipes)
- Crude protein extraction buffer (see Recipes)
Equipment
- Pipettes
- pH meter
- Weighing balance
- Laminar flow hood
- Stirring bar
- Magnetic stirrer (VWR, model: 200 Mini-stirrer )
- Vortex (Fisher Scientific, model: Vortex-Genie 2 )
- Scintillation counter (Beckman Coulter, model: LS-6500 )
- Centrifuge (Beckman Coulter, model: Microfuge® 22R , catalog number: 368831)
- Spectrophotometer (Bio-Rad Laboratories, model: SmartSpec Plus, catalog number: 1702525 )
Note: This product has been discontinued. - Autoclave
Procedure
- Preparation of callus cells and plant seedlings
- Callus cells
- Put the Arabidopsis seeds in seed sterilization solution (see Recipes) for 15-20 min. Discard the sterilization solution and resuspend the seeds with sterilized ddH2O in a laminar flow hood. Let the seeds settle down and discard the solution. Repeat the washing step 5 times.
- Sow the surface-sterilized seeds in a row on callus induction medium (see Recipes) in a laminar flow hood, seal the Petri dishes with Parafilm and incubate the Petri dishes at 4 °C in the dark for 2 days.
- Place the Petri dishes in a horizontal position and let the seeds germinate and grow under 16 h light/8 h dark cycle (lighting provided by fluorescent bulbs giving an average light intensity of ~150 µmol/m2/sec) at 22 °C for 3 weeks.
- Transfer induced callus (Figure 1) to new callus induction medium in a laminar flow hood every 3 weeks until the transport assay (see Note 1).
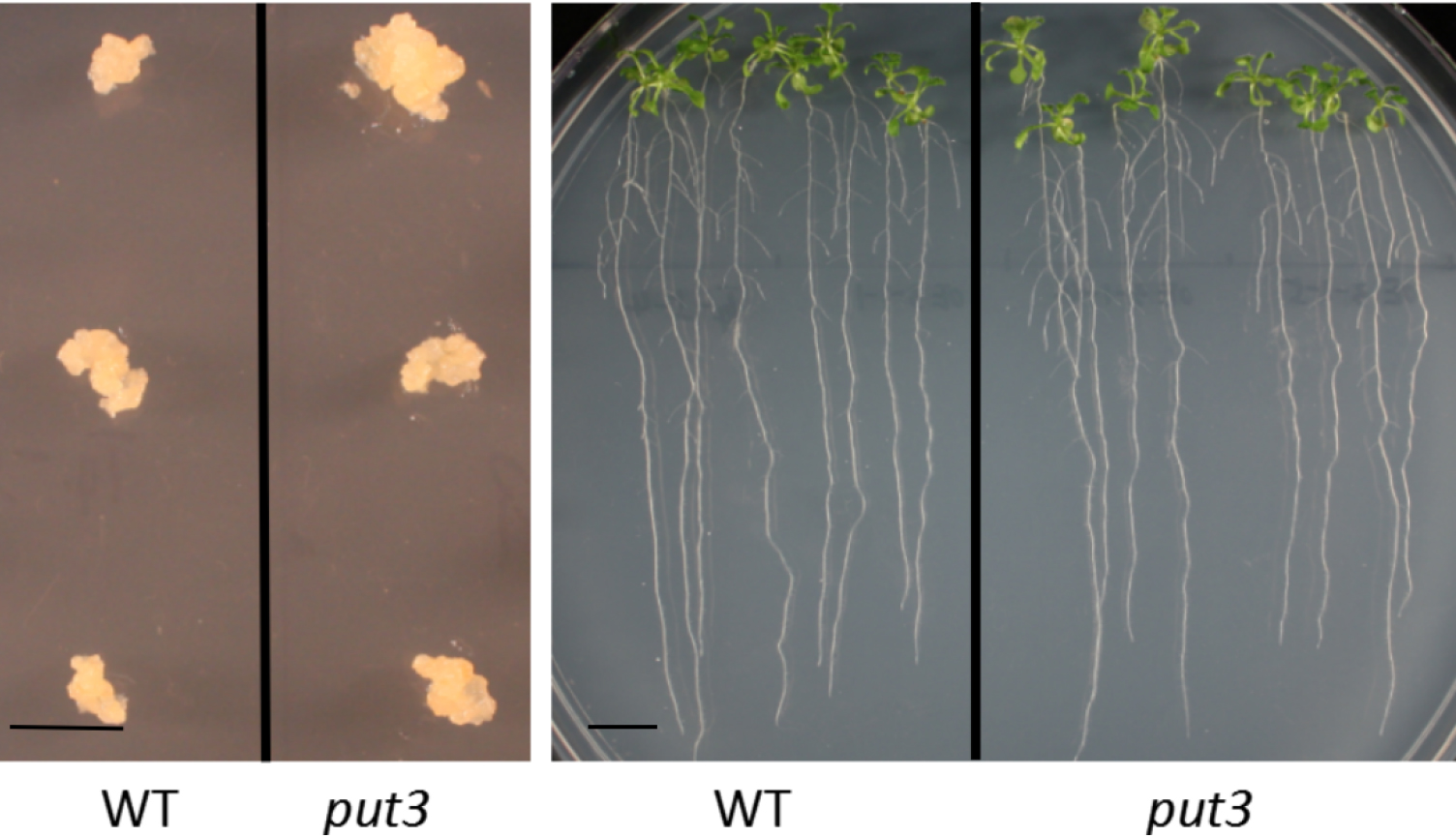
Figure 1. 3-week-old induced callus cells and 2-week-old Arabidopsis seedlings. Bars = 1 cm. - Plant seedlings
- Put the Arabidopsis seeds in seed sterilization solution for 15-20 min. Discard the sterilization solution and resuspend the seeds with sterilized ddH2O in a laminar flow hood. Let the seeds settle down and discard the solution. Repeat the washing step 5 times.
- Sow the surface-sterilized seeds in a row on ½ MS medium (see Recipes) in a laminar flow hood, seal the Petri dishes with Parafilm and incubate the Petri dishes at 4 °C in the dark for 2 days.
- Place the Petri dish in a vertical position and let the seeds germinate and grow under 16 h light/8 h dark cycle (lighting provided by fluorescent bulbs giving an average light intensity of ~150 µmol/m2/sec) at 22 °C for 2 weeks (Figure 1).
- Generation of standard curve
- Standard curve for 14C-labeled PQ
- Make a 50x and 500x dilution of 10 µl solution 2 (see Recipes) with ddH2O.
- In 1 ml of the ScintiVerseTM BD Cocktail, add 0 µl, 25 µl, 125 µl and 250 µl of 500x diluted solution to make 14C-labeled PQ solutions with 0 nCi, 0.1 nCi, 0.5 nCi, 1 nCi radioactivity, and add 125 µl and 250 µl of 50x diluted solution to make 14C-labeled PQ with 5 nCi and 10 nCi radioactivity, respectively.
- Measure the radioactivity by using Beckman LS-6500 scintillation counter to generate a standard curve for 14C-labeled PQ.
- Perform 3 sets of this test to ensure accuracy of the standard curve (Figure 2).
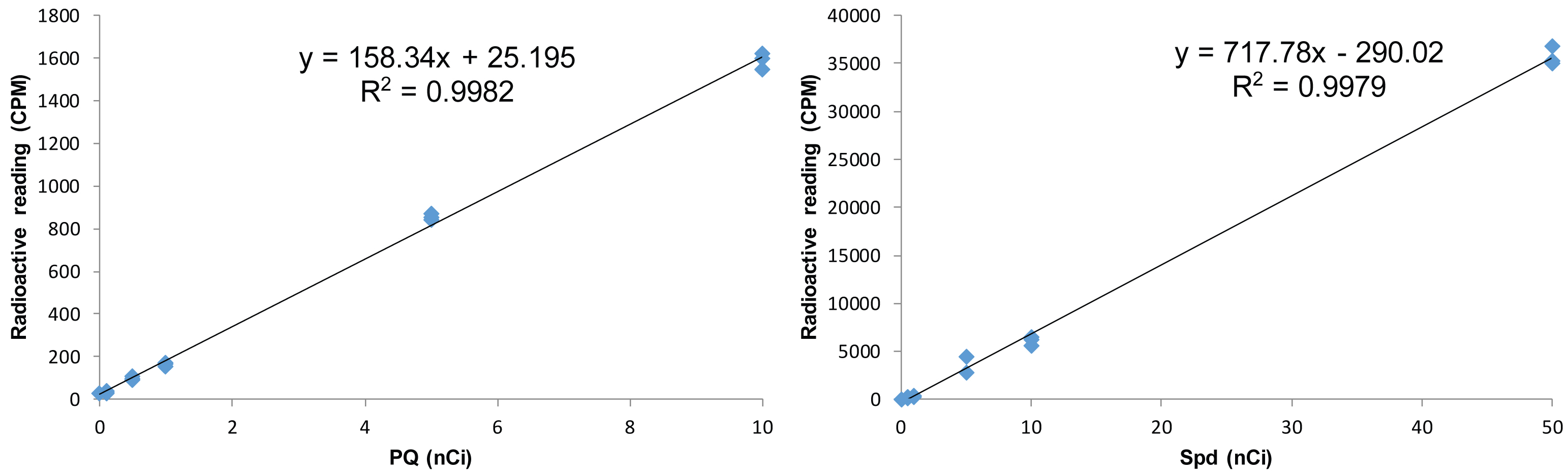
Figure 2. Representative of standard curves for radioactive labeled PQ and Spd
- Make a 50x and 500x dilution of 10 µl solution 2 (see Recipes) with ddH2O.
- Standard curve for 3H-labeled Spd
- Make a 100x dilution of 10 µl solution 3 (see Recipes) with ddH2O.
- In 1 ml of the ScintiVerseTM BD Cocktail, add 0 µl, 2.5 µl, 5 µl, 25 µl, 50 µl and 250 µl 100x diluted solution to make 3H-labeled Spd solutions with 0 nCi, 0.5 nCi, 1 nCi, 5 nCi, 10 nCi and 50 nCi radioactivity, respectively.
- Measure the radioactivity by using Beckman LS-6500 scintillation counter to generate a standard curve for 3H-labeled Spd.
- Perform 3 sets of this test to ensure accuracy of the standard curve (Figure 2).
- Standard curve for 14C-labeled PQ
- Transport assays
- Transport assays with callus (Table 1)
Table 1. Designed temperatures and Spd or PQ concentrations used in transport assay of callus cells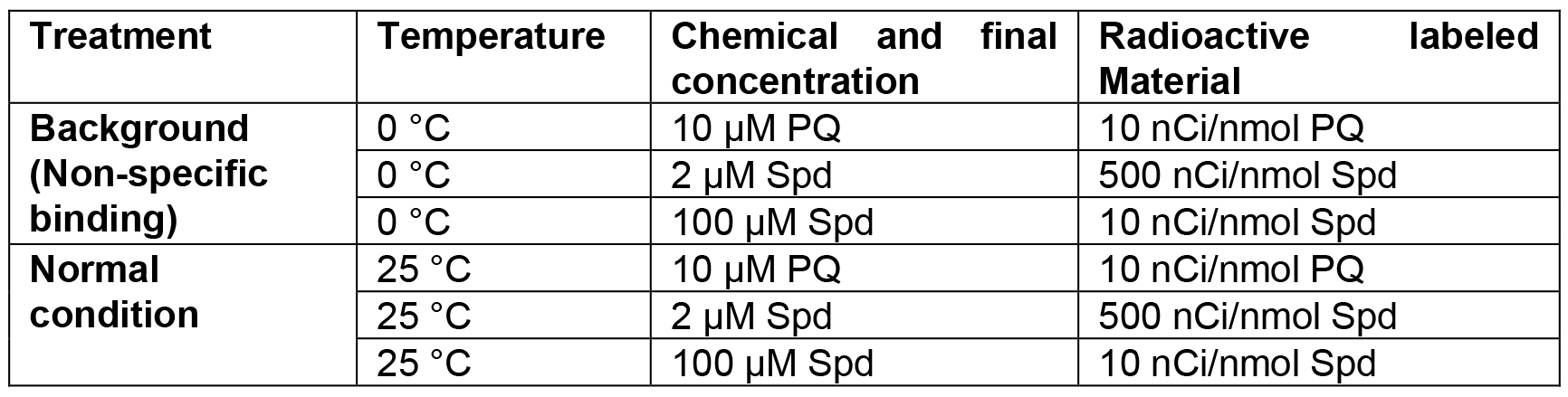
- Transfer 100 mg fresh callus cells into a 1.5 ml Eppendorf centrifuge tube containing 950 µl liquid callus-inducing medium and vortex for 30 sec.
- Pre-treat samples at designed temperature for 30 min to reach equilibrium (Shen et al., 2016).
- Add 50 µl solution 2 (final PQ concentration at 10 µM), solution 3 (final Spd concentration at 2 µM) or solution 4 (final Spd concentration at 100 µM) (see Recipes) into the tubes according to the treatment conditions (see Note 2).
- Briefly vortex for mixing and return the tubes back to the designed temperature.
- Incubate for 1 h, gently invert each tube by hand every 15 min to resuspend the cells and ensure the equilibrium of PQ or Spd in the solution.
- Place tubes on ice immediately.
- Let the callus cells settle down and remove the PQ- or Spd-containing liquid callus inducing medium by using pipette.
- Wash callus cells five times on ice each with 1.5 ml pre-chilled liquid callus inducing medium. For each treatment, three replicate samples should be used at the same time.
- Transfer 100 mg fresh callus cells into a 1.5 ml Eppendorf centrifuge tube containing 950 µl liquid callus-inducing medium and vortex for 30 sec.
- Transport assays with seedlings (Table 2)
Table 2. Designed temperatures and Spd or PQ concentrations used in transport assay of plant seedlings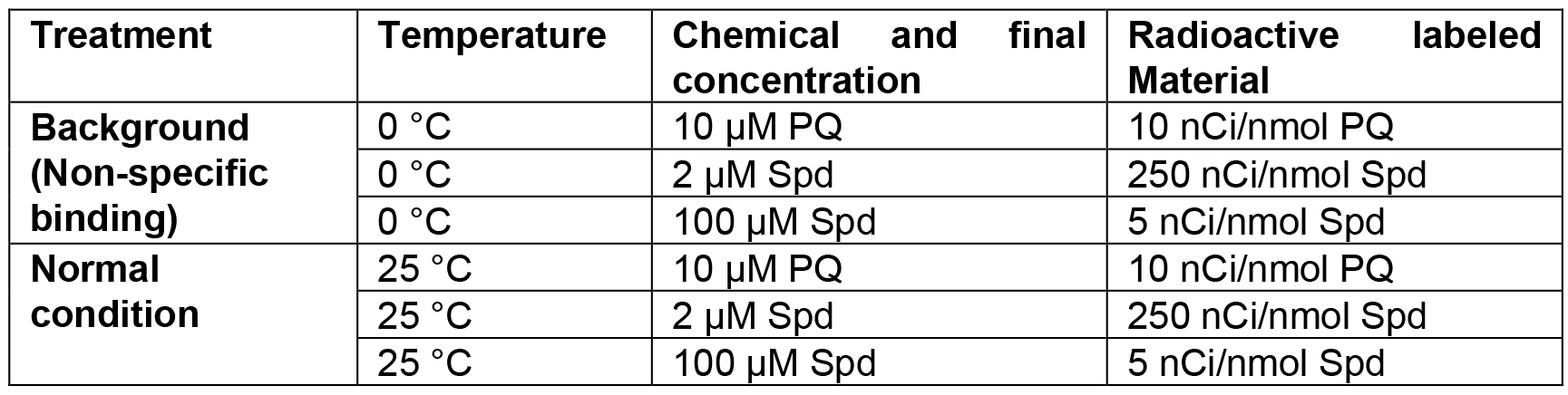
- Transfer 20 seedlings of both wild type Col-0 and put3 mutant line with intact roots onto a filter paper saturated with 5 ml liquid ½ MS medium (see Recipes), and pre-incubate the seedlings at the designed temperature for 30 min (see Note 3).
- Mix 250 µl solution 2 (final PQ concentration at 10 µM), solution 5 (final Spd concentration at 2 µM) (see Recipes), or solution 6 (final Spd concentration at 100 µM) (see Recipes) with 4.75 ml ½ MS medium to reach the final concentrations.
- Saturate another piece of filter paper with the radioactive solution made in step C2b and pre-incubate it at the designed temperature for 30 min.
- Transfer pre-incubated seedlings in the step C2a one by one to the filter paper containing radioactive-labeled PQ or Spd and pre-incubate at the same temperature as stated in the step C2c. Carefully arrange the seedlings to make sure all roots are directly in contact with the filter paper.
- Incubate at designed temperature for 1 h.
- Cut off roots of the seedlings from each treatment.
- Transfer the roots from each treatment into a 1.5 ml Eppendorf centrifuge tube.
- Wash the roots 5 times on ice by using 1.5 ml pre-chilled liquid ½ MS medium. For each treatment, three replicate samples should be used at the same time.
- Transport assays with callus (Table 1)
- Extraction of intracellular contents
- Grind washed callus cells or seedling roots by using small blue pestle in 1 ml (for callus cells) or 500 µl (for seedling roots) crude protein extracting buffer (see Recipes) on ice until no clumps remain to release all intracellular contents. The solution should be homogenized to an even mixture that can be pipetted without clogs.
- Centrifuge the mixture at 15,500 x g for 20 min at 4 °C.
- Transfer supernatant into a new tube for radioactive and protein concentration measurements.
- Measure the UV absorbance of 300 µl (for callus cells) or 100 µl (for seedling roots) of the supernatant of each sample by using Bio-Rad SmartSpec Plus Spectrophotometer. The total protein concentration is then calculated based on the UV absorbance using the equation:
Protein concentration (mg/ml) = (1.55 x A280) - (0.76 x A260)
- Grind washed callus cells or seedling roots by using small blue pestle in 1 ml (for callus cells) or 500 µl (for seedling roots) crude protein extracting buffer (see Recipes) on ice until no clumps remain to release all intracellular contents. The solution should be homogenized to an even mixture that can be pipetted without clogs.
- Radioactive measurement
- Add 300 µl (for callus cells) or 100 µl (for seedling roots) of the supernatant of each sample into 1 ml ScintiVerseTM BD Cocktail solution.
- Measure the radioactivity of each sample by using Beckman LS-6500 scintillation counter.
- Add 300 µl (for callus cells) or 100 µl (for seedling roots) of the supernatant of each sample into 1 ml ScintiVerseTM BD Cocktail solution.
Data analysis
The radioactivity (nCi) of each sample was calculated based on the scintillation counter reading (CPM) and the equation of the corresponding standard curve. The total amount of transported PQ/Spd in each sample was then calculated based on the radioactivity and the used nCi per nmol in the transport assay solution. The total protein of each sample was calculated based on the protein concentration and the volume. The PQ/Spd transport rates were presented in the unit of µmole PQ/Spd per gram protein per hour (µmole g-1 h-1). The PQ/Spd transport rate at 0 °C of each sample was considered as non-specific binding of PQ/Spd, and therefore was subtracted from the PQ/Spd transport rates at 25 °C to calculate the adjusted and accurate PQ/Spd transport rates at normal condition. Readers are referred to Shen et al. (2016) for examples of data graphs.
Notes
- We maintained the callus cells by transferring fresh and fast-growing callus cells to a new callus induction medium every 3 weeks. By judging the color and growth of the callus cells, it is easy to select the callus cells for transfer. Fresh calli usually show light color whereas relative old calli show dark color.
- 2 µM Spd was designed to test if the transporter is a high-affinity PA transporter whereas 100 µM Spd was designed to test if other transporters are also working at high Spd concentrations. In our test, the callus cells and seedlings of the put3 mutant showed approximately 20-fold and 4-fold reduction, respectively, on 2 µM Spd uptake rate compared with the wild type. However, when a higher concentration (100 µM) of Spd was added into the medium, the transport rate was comparable in both put3 mutant and wild type. This might suggest that PUT3 protein is a high-affinity PA transporter and some low-affinity PA transporters may start to work at high PA concentrations.
- In the transport assay with seedlings, we used 150 x 15 mm Petri dish and put a filter paper fitting into the Petri dish. Seedlings were arranged relatively separate.
Recipes
- Seed sterilization solution
25% Clorox Bleach
0.05% Triton X-100
Store at room temperature - 2,000x 2,4-D (9.05 mM)
26 mg (2,4-dichlorophenoxy) acetic acid sodium salt monohydrate
Add ddH2O to 11 ml and sterilize by filtration through a syringe filter (0.2 µm)
Store at -20 °C - 2,000x kinetin (1.86 mM)
10 mg kinetin
1 ml 1 N NaOH
Add ddH2O to 25 ml and sterilize by filtration through a syringe filter (0.2 µm)
Store at -20 °C - Callus induction solid medium
4.33 g/L Murashige & Skoog basal salts
3% sucrose
Adjust pH to 5.7 with 0.1 N NaOH and add agar to 0.7% (w/v)
Autoclave
Cool medium to ~60 °C
Add 2,4-D to a final concentration at 4.52 µM
Add Kinetin to a final concentration at 0.93 µM
Store at room temperature - Callus induction liquid medium
4.33 g/L Murashige & Skoog basal salts
3% sucrose
Adjust pH with 0.1 N NaOH to 5.7 and autoclave
Cool medium to ~60 °C
Add 2,4-D to a final concentration at 4.52 µM
Add Kinetin to a final concentration at 0.93 µM
Store at room temperature - ½ MS solid medium
2.17 g/L Murashige & Skoog basal salts
1.5% sucrose
Adjust pH to 5.7 with 0.1 N NaOH and add agar to 1.2% (w/v)
Autoclave
Store at room temperature - ½ MS liquid medium
2.17 g/L Murashige & Skoog basal salts
1.5% sucrose
Adjust pH with 0.1 N NaOH to 5.7 and autoclave
Store at room temperature - 200 µM non-14C-labeled PQ solution
5.14 mg methyl viologen dichloride hydrate
100 ml ddH2O
Store at -20 °C - 2.04 mM non-3H-labeled Spd solution
3.20 µl spermidine
10 ml ddH2O
Store at -20 °C - 40.37 µM non-3H-labeled Spd solution
198 µl 2.04 mM spermidine
9.8 ml ddH2O
Store at -20 °C - Solution 1: 14C-labeled PQ solution (200 µM, 32.3 nCi/nmol, 15.55 ml)
0.8 mg paraquat-methyl-C14 dichloride
15.55 ml ddH2O
Store at -20 °C - Solution 2: 14C-labeled PQ solution (200 µM, 10 nCi/nmol, 1 ml)
309.6 µl solution 1
690.4 µl 200 µM non-14C-labeled PQ solution
Store at -20 °C - Solution 3: 3H-labeled Spd solution (40 µM, 500 nCi/nmol, 1 ml)
20 µl spermidine trihydrochloride, [terminal methylenes-3H (N)] - solution
980 µl 40.37 µM non-3H-labeled Spd solution
Store at -20 °C - Solution 4: 3H-labeled Spd solution (2 mM, 10 nCi/nmol, 1 ml)
20 µl spermidine trihydrochloride, [terminal methylenes-3H (N)] - solution
980 µl 2.04 mM non-3H-labeled Spd solution
Store at -20 °C - Solution 5: 3H-labeled Spd solution (40 µM, 250 nCi/nmol, 1 ml)
500 µl solution 3
500 µl 40 µM non-3H-labeled Spd solution
Store at -20 °C - Solution 6: 3H-labeled Spd solution (2 mM, 5 nCi/nmol, 1 ml)
500 µl solution 4
500 µl 2 mM non-3H-labeled Spd solution
Store at -20 °C - 1 M Tris-HCl, pH 7.5
121.1 g Trizma® base
800 ml ddH2O
Adjust pH to 7.5 with concentrated HCl
Add ddH2O to 1,000 ml, autoclave
Store at room temperature - 0.5 M EDTA, pH 8.0
186.1 g ethylenediaminetetraacetic acid disodium salt dihydrate
800 ml ddH2O
Adjust pH to 8.0 with NaOH (about 20 g)
Add ddH2O to 1,000 ml, autoclave
Store at room temperature - Crude protein extraction buffer
50 mM Tris-HCl (pH 7.5)
0.1 mM EDTA
1 mM β-mercaptoethanol
Make fresh solution and store at room temperature
Acknowledgments
This work was partly supported by the US Department of Agriculture National Research Initiative competitive grant 2007-35100-18378 to H.S. and by the National Natural Science Foundation of China (Grant # 31328004) to H.S.
References
- Gill, S. S. and Tuteja, N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 5(1): 26-33.
- Li, J., Mu, J., Bai, J., Fu, F., Zou, T., An, F., Zhang, J., Jing, H., Wang, Q., Li, Z., Yang, S. and Zuo, J. (2013). Paraquat Resistant1, a Golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiol 162(1): 470-483.
- Mulangi, V., Chibucos, M. C., Phuntumart, V. and Morris, P. F. (2012). Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236(4): 1261-1273.
- Shen, Y., Ruan, Q., Chai, H., Yuan, Y., Yang, W., Chen, J., Xin, Z. and Shi, H. (2016). The Arabidopsis polyamine transporter LHR1/PUT3 modulates heat responsive gene expression by enhancing mRNA stability. Plant J 88(6): 1006-1021.
- Uemura, T., Kashiwagi, K. and Igarashi, K. (2007). Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J Biol Chem 282(10): 7733-7741.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chai, H., Shen, Y. and Shi, H. (2017). Polyamine and Paraquat Transport Assays in Arabidopsis Seedling and Callus. Bio-protocol 7(15): e2421. DOI: 10.21769/BioProtoc.2421.
Category
Plant Science > Plant physiology > Tissue analysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










