- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differentiation of Human Induced Pluripotent Stem Cells (iPS Cells) and Embryonic Stem Cells (ES Cells) into Dendritic Cell (DC) Subsets
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2419 Views: 16507
Reviewed by: Xiao LiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Matthijs Snelders [...] Jeroen Essers
Mar 5, 2025 3912 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2411 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 210 Views
Abstract
Induced pluripotent stem cells (iPS cells) are engineered stem cells, which exhibit properties very similar to embryonic stem cells (ES cells; Takahashi and Yamanaka, 2016). Both iPS cells and ES cells have an extraordinary self-renewal capacity and can differentiate into all cell types of our body, including hematopoietic stem/progenitor cells and dendritic cells (DC) derived thereof. This makes iPS cells particularly well suited for studying molecular mechanisms of diseases, drug discovery and regenerative therapy (Grskovic et al., 2011; Bellin et al., 2012; Robinton and Daley, 2012).
DC are the major antigen presenting cells of the immune system and thus they are key players in modulating and directing immune responses (Merad et al., 2013). DC patrol peripheral and interface tissues (e.g., lung, intestine and skin) to detect invading pathogens, and upon activation they migrate to lymph nodes to activate and prime lymphocytes.
DC comprise a phenotypically heterogeneous family with functionally specialized subsets (Schlitzer and Ginhoux, 2014). Generally, classical DC (cDC) and plasmacytoid DC (pDC) are distinguished, exhibiting a classical and plasma cell-like DC morphology, respectively. cDC recognize a multitude of pathogens and secrete proinflammatory cytokines upon activation, while pDC are specialized to detect intracellular pathogens and secrete type I interferons (Merad et al., 2013; Schlitzer and Ginhoux, 2014). cDC are further divided into cross-presenting cDC1 and conventional cDC2, in the human system referred to as CD141+ Clec9a+ cDC1 and CD1c+ CD14- cDC2. Human pDC are characterized as CD303+ CD304+ (Jongbloed et al., 2010; Joffre et al., 2012; Swiecki and Colonna, 2015).
To investigate subset specification and function of human DC, we established a protocol to generate cDC1, cDC2 and pDC in vitro from human iPS cells (or ES cells) (Sontag et al., 2017). Therefore, we differentiated iPS cells (or ES cells), via mesoderm commitment and hemato-endothelial specification, into CD43+ CD31+ hematopoietic progenitors. Subsequently, those were seeded onto inactivated OP9 stromal cells with FLT3L, SCF, GM-CSF and IL-4 or FLT3L, SCF and GM-CSF to specify cDC1 and cDC2, or cDC1 and pDC, respectively.
Background
DC and their development have mostly been studied in mice (Belz and Nutt, 2012; Merad et al., 2013; Schlitzer and Ginhoux, 2014). Owing to their rarity in non-lymphoid tissues and the restricted access to human lymphoid tissues, studying DC in humans is challenging (Jongbloed et al., 2010; Villadangos and Shortman, 2010). Yet, understanding developmental pathways, origins and mechanisms during DC subset specification is important in order to generate autologous DC subsets in vitro for therapeutic applications (e.g., anti-tumor agents). iPS cells (and ES cells) with their unlimited self-renewal and differentiation potential raise hopes that DC can be generated and modified (e.g., loaded with patient and disease specific antigens) in high numbers and quality in vitro to study DC development and function and to improve DC immunotherapies (Sontag et al., 2017).
Several groups have generated monocyte derived DC from iPS cells (and ES cells) with conventional granulocyte macrophage colony stimulating factor (GM-CSF)/interleukin 4 (IL-4) protocols, but such DC represent inflammatory DC and are not subset specific (Senju et al., 2007; Tseng et al., 2009; Choi et al., 2011; Senju et al., 2011; Belz and Nutt, 2012; Rossi et al., 2012; Yanagimachi et al., 2013; Li et al., 2014). From mouse studies, it is known that fms-like tyrosine kinase ligand (FLT3L) is the key cytokine for DC development and that the combination of FLT3L and GM-CSF signaling specifies DC subsets in vivo (Gilliet et al., 2002; Schmid et al., 2010). Recently, human cDC1, cDC2 and pDC were generated from cord blood (CB) with FLT3L, stem cell factor (SCF) and GM-CSF on MS5 stromal cells (Lee et al., 2015). Additionally, Poulin et al. (2010) reported on the generation of cDC1 from CB with FLT3L, SCF, GM-CSF and IL-4 in a feeder-free environment (Poulin et al., 2010). In contrast, Silk et al. (2012) described the differentiation of cDC1 in a feeder-free GM-CSF/IL-4 system (Silk et al., 2012). However, recent genome wide transcriptional profiling studies highlight the impact of microenvironmental cues during DC development, indicating that feeder support is important (Lundberg et al., 2013). Therefore, we used FLT3L, SCF, GM-CSF and IL-4 (referred to as FSG4) and FLT3L, SCF and GM-CSF (referred to as FSG) in combination with OP9 stromal cells to generate cDC1, cDC2 and pDC from iPS cell (or ES cell) derived hematopoietic progenitors.
Here we describe a two-step protocol: First, human iPS cells (or ES cells) are differentiated into hematopoietic progenitors (adapted from Kennedy et al., 2012). Second, these hematopoietic stem/progenitor cells are then further differentiated into cDC1, cDC2 and pDC (Sontag et al., 2017).
Materials and Reagents
- 6-well tissue culture plates (TPP Techno Plastic Products, catalog number: 92006 )
- 10 cm microbiology grade Petri dishes (SARSTEDT, catalog number: 82.1473.001 )
- 50 ml Falcon tubes (Corning, Falcon®, catalog number: 352070 )
- 70 µm cell strainer (Greiner Bio One International, catalog number: 542070 )
- 40 µm cell strainer (Greiner Bio One International, catalog number: 542040 )
- Top bottle filter (TPP Techno Plastic Products, catalog number: 99505 )
- 5 ml Serological pipettes (Corning, Falcon®, catalog number: 357543 )
- 10 ml Serological pipettes (Corning, Falcon®, catalog number: 357551 )
- 25 ml Serological pipettes (Corning, Falcon®, catalog number: 357525 )
- Pipette tips 10 µl (STARLAB INTERNATIONAL, catalog number: S1180-3810 )
- Pipette tips 20 µl (STARLAB INTERNATIONAL, catalog number: S1120-1810 )
- Pipette tips 200 µl (STARLAB INTERNATIONAL, catalog number: S1120-8810 )
- Pipette tips 1,000 µl (STARLAB INTERNATIONAL, catalog number: S1126-7810 )
- Collagenase IV (Thermo Fisher Scientific, GibcoTM, catalog number: 17104019 )
- Knockout-Dulbecco’s modified Eagle medium (KO-DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 10829018 )
- Gelatin (Sigma-Aldrich, catalog number: G1890 )
- 1-Thiogylcerol (Sigma-Aldrich, catalog number: M1753 )
- L-ascorbic acid (Sigma-Aldrich, catalog number: A4403 )
- Bovine serum albumin (BSA) low endotoxin (PAA, catalog number: K31-011 )
- 1x Dulbecco’s phosphate buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14190094 )
- HumanKine bone morphogenic protein 4 (BMP4) research grade (Miltenyi Biotec, catalog number: 130-110-921 )
- Recombinant human basic fibroblast growth factor (bFGF) (PeproTech, catalog number: 100-18B )
- Recombinant human FLT3L (PeproTech, catalog number: 300-19 )
- Recombinant human GM-CSF (PeproTech, catalog number: 300-03 )
- Recombinant human insulin-like growth factor 1 (IGF1) (PeproTech, catalog number: 100-11 )
- Human interleukin 3 (IL-3) research grade (Miltenyi Biotec, catalog number: 130-094-193 )
- Human IL-4 research grade (Miltenyi Biotec, catalog number: 130-095-373 )
- Human SCF research grade (Miltenyi Biotec, catalog number: 130-096-693 )
- Human thrombopoietin (TPO) research grade (Miltenyi Biotec, catalog number: 130-094-013 )
- Recombinant human vascular endothelial growth factor (VEGF) (PeproTech, catalog number: 100-20 )
- 1x StemPro-34 SFM (Thermo Fisher Scientific, catalog number: 10639011 )
- Penicillin-streptomycin 10,000 U/ml (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- L-glutamine 200 mM (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Hyper-interleukin 6 (IL-6) (kindly provided by S. Rose John, Institute of Biochemistry, Medical Faculty, Christian-Albrechts-University, Kiel, Germany, [Fischer et al., 1997], see also Notes)
- α-Minimal essential medium (α-MEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 12571063 )
- β-mercaptoethanol (50 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 31350010 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10500064 )
- Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093 )
- Collagenase IV solution (1 mg/ml) (see Recipes)
- 0.1% gelatin solution (see Recipes)
- 1-Thioglycerol stock solution (100 mM) (see Recipes)
- L-ascorbic acid stock solution (50 mg/ml) (see Recipes)
- 0.1% BSA solution (see Recipes)
- BMP4 stock solution (25 µg/ml) (see Recipes)
- bFGF stock solution (100 µg/ml) (see Recipes)
- FLT3L stock solution (25 µg/ml) (see Recipes)
- GM-CSF stock solution (100 µg/ml) (see Recipes)
- IGF1 stock solution (40 µg/ml) (see Recipes)
- IL-3 stock solution (150 µg/ml) (see Recipes)
- IL-4 stock solution (20 µg/ml) (see Recipes)
- SCF stock solution (100 µg/ml) (see Recipes)
- TPO stock solution (20 µg/ml) (see Recipes)
- VEGF stock solution (100 µg/ml) (see Recipes)
- Hematopoietic progenitor basal differentiation medium (see Recipes)
- Induction medium d0 (see Recipes)
- Induction medium d1 (see Recipes)
- Induction medium d2 (see Recipes)
- Induction medium d4 (see Recipes)
- Induction medium d6 (see Recipes)
- OP9 culture medium (see Recipes)
- DC differentiation basal medium (see Recipes)
- DC differentiation FSG4 medium (see Recipes)
- DC differentiation FSG medium (see Recipes)
Equipment
- 1,000 µl pipette (Gilson, catalog number: F123602 )
- 200 µl pipette (Gilson, catalog number: F123601 )
- 20 µl pipette (Gilson, catalog number: F123600 )
- 10 µl pipette (Gilson, catalog number: F144802 )
- Pipetboy (INTEGRA Biosciences, catalog number: 155000 )
- Water bath (JULABO, model: SW22 )
- Inverted light microscope (Leica Microsystems, model: Leica DM IL LED )
- Autoclave
- Centrifuge (Thermo Fisher Scientific, model: HeraeusTM Multifuge 3 L )
- Flow hood (Heraeus)
- Automatic CO2 incubator with nitrogen supply and O2 sensor (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM 240i , catalog number: 51026331)
- Vacuum pump (INTEGRA Biosciences, catalog number: 158300 )
- Standard fridge
- Standard non-defrosting freezer
Procedure
- The differentiation of iPS cells (or ES cells) into hematopoietic progenitors is summarized in Figure 1.
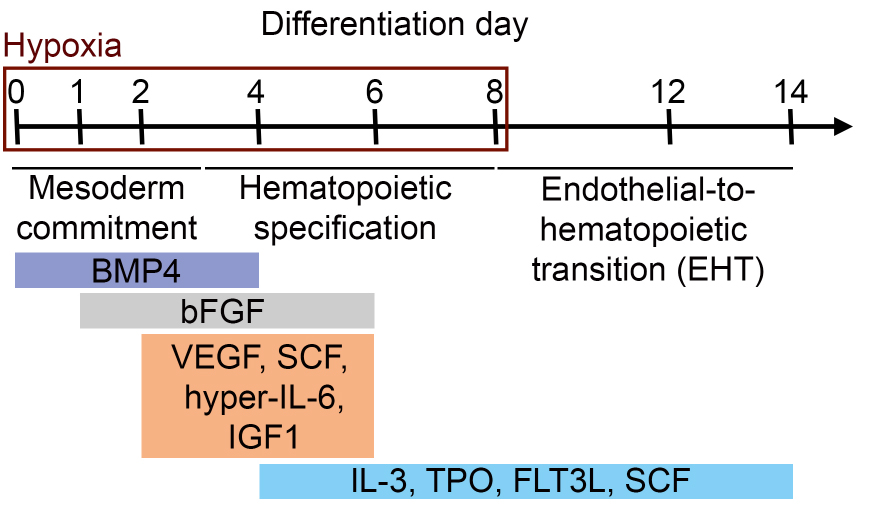
Figure 1. Schematic representation of the differentiation protocol of iPS cells (or ES cells) into hematopoietic progenitors. Differentiation time and corresponding differentiation steps and cytokine compositions (in colored boxes) are shown.- To differentiate iPS cells (or ES cells) into hematopoietic progenitors, prewarm required amounts of collagenase IV solution (1 mg/ml, 37 °C, see Recipes) and KO-DMEM (room temperature, RT) before starting the experiment. Prepare induction medium d0 (see Recipes) and prewarm at RT.
- Harvest iPS cells (or ES cells) when they reached 70-80% confluence. Therefore, aspirate medium and add collagenase IV solution to the cells. The volume depends on the culture dish used. Make sure to completely cover the cells with collagenase IV solution (e.g., 1 ml per well of a 6-well plate, 4 ml per 10 cm dish).
- Incubate cells with collagenase IV at 37 °C in a normoxic incubator with 5% CO2 for 40-60 min. Check if colony edges start to roll up. Only if colony edges have rolled up from all sites and colonies are only loosely adherent to the plate proceed with step A4. If necessary, incubate 10-15 min longer but do not exceed 80 min collagenase IV treatment because this compromises cell viability.
- Dilute collagenase IV with KO-DMEM in a ratio 1/3, meaning add 2 ml KO-DMEM for each ml collagenase IV solution. Use a 10 ml pipette to rinse off colonies, resuspend and collect them in 50 ml Falcon tubes.
- If iPS cells (or ES cells) were cultured on mouse embryonic fibroblasts (MEF), separate iPS cell (or ES cell) clusters and single MEF by gravity for 10 min at RT. Large iPS cell (or ES cell) clusters will settle to the bottom, while single MEF will remain in suspension. Aspirate 80% of the supernatant to remove MEF. If iPS cells (or ES cells) were cultured feeder-free, directly continue with step A6.
- Collect iPS cell (or ES cell) clusters by centrifugation at 212 x g for 4 min. If not stated otherwise, all centrifugation steps are performed at RT. Aspirate supernatant and resuspend pellet in 1 ml induction medium d0. Pipette up and down with 1 ml pipette to break up clusters. Clusters should contain 50-100 cells (Figure 2). Therefore, transfer a drop of the cell suspension onto a microbiology grade Petri dish and check cluster size by microscopy.
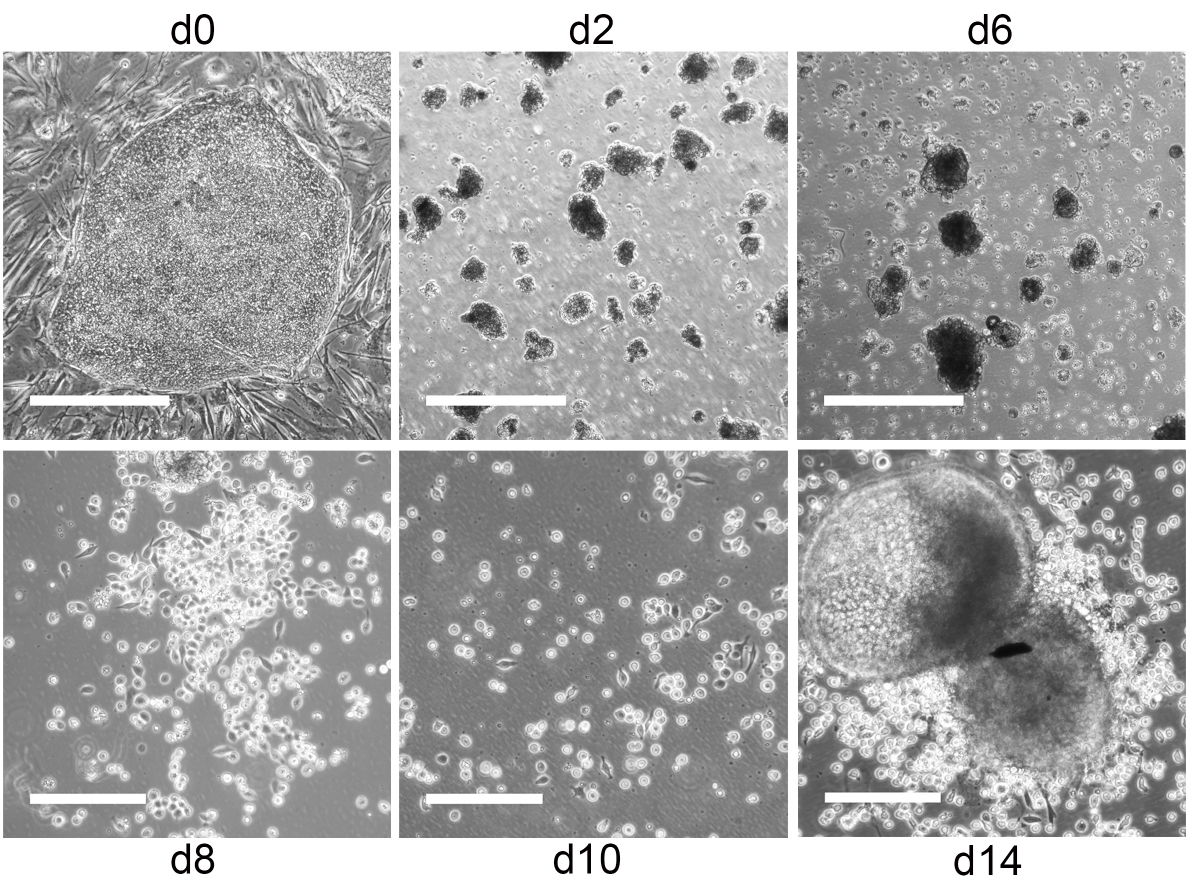
Figure 2. From iPS cells to hematopoietic progenitors. Representative phase contrast images of iPS cells during differentiation into hematopoietic progenitors. Undifferentiated iPS cell colonies on day 0 and small EB with some cell debris on day 2 and 6. Scale bars = 500 µm. Hemato-endothelial patches and hematopoietic progenitors on day 8, 10 and 14. Scale bars = 200 µm. - If cluster size is appropriate, pass cell suspension through a 70 µm cell strainer to remove remaining larger clusters. Fill up flow-through to the final volume with induction medium d0 and transfer to microbiology grade Petri dishes to allow embryoid bodies (EB) to form. We usually use 10 ml induction medium d0 for two 6-well plates with iPS cells (or ES cells).
- Place cells in a 37 °C, 5% CO2 incubator and set O2 regulator of the automatic incubator to 5%. Thus, O2 levels will slowly decrease to 5%.
- On day 1 of differentiation (24 h after EB formation), many cells (approx. 40-50%) will have died. Prepare induction medium d1 (see Recipes) and prewarm at RT. Dilute old medium (induction medium d0) with fresh medium (induction medium d1) in a ratio of 1/2. Calculate volume of bFGF in induction medium d1 according to total volume (old medium + fresh medium).
Example: 10 ml induction medium d0 + 5 ml induction medium d1 medium containing 1.5 µl bFGF and 1.6 µl BMP4. Final bFGF concentration should be 10 ng/ml. Continue culture under hypoxic conditions (5% O2, 5% CO2). - On day 2 of differentiation, there will be a lot of cell debris visible (Figure 2). Collect EB in 50 ml Falcon tubes. Despite the use of microbiology grade Petri dishes, some EB loosely adhere to the dish. Rinse loosely adherent EB off the dishes twice with 1x PBS (prewarmed to RT) and add to EB suspension. Centrifuge EB at 136 x g for 4 min. Aspirate supernatant, gently resuspend pellet (if necessary with 1 ml pipette to break up aggregated EB) in induction medium d2 (prewarmed to RT, see Recipes) and transfer to new microbiology grade Petri dishes. Use the same volume as on day 0 (see step A7). Continue culture under hypoxic conditions (5% O2, 5% CO2).
- On day 4 of differentiation, proceed as in step A10. Finally, resuspend EB in induction medium d4 (prewarmed to RT, see Recipes) and transfer to new microbiology grade Petri dishes. Use the same volume as on day 0 (see step A7). Continue culture under hypoxic conditions (5% O2, 5% CO2).
- On day 6 of differentiation, proceed as in step A10. Finally, resuspend EB in induction medium d6 (prewarmed to RT, see Recipes) and transfer to tissue culture plastic (TCP) dishes coated with 0.1% gelatin (see Recipes). Use the same volume as on day 0 (see step A7). Continue culture under hypoxic conditions (5% O2, 5% CO2).
- On day 8 of differentiation, most EB are adherent on TCP. Collect medium and centrifuge at 136 x g for 4 min. Aspirate supernatant, resuspend in induction medium d6 (prewarmed to RT) and transfer carefully back to the TCP dish. Use the same volume as on day 0 (see step A7). From now on, avoid detaching EB from the dish. Transfer cells to 37 °C, 5% CO2 and normoxic incubator.
- On day 10 and 12 of differentiation, medium change is performed as in step A13.
- Depending on the iPS cell (or ES cell) clone used hematopoietic progenitors will start to bud off from adherent or loosely adherent EB between day 6-14 (iPS cells) or day 21-28 (ES cells) of differentiation, respectively (Figure 2, Video 1). iPS cell (or ES cell) derived hematopoietic progenitors are CD43+ (Figure 3) and used for DC differentiation between day 10-14 (iPS cells) and day 24-28 (ES cells).
Video 1. Emergence of iPS cell (or ES cell) derived hematopoietic progenitors. Representative video of iPS cell derived EB on day 6 of differentiation.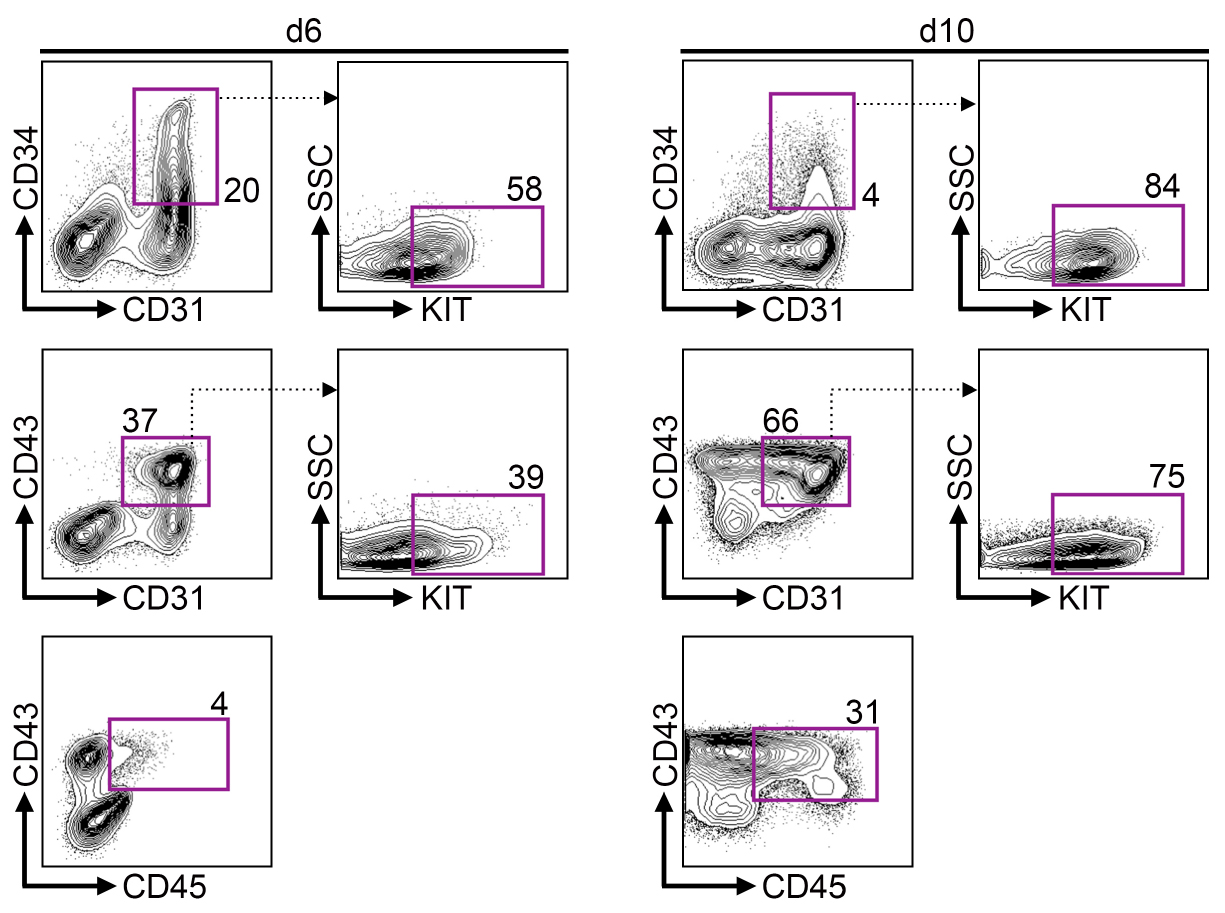
Figure 3. iPS cells develop via hemogenic endothelium into hematopoietic progenitors. Representative flow cytometry data on day 6 and 10 of differentiation. Upper row shows gating for CD34+ CD31+ hemogenic endothelium population. Center row shows gating for CD43+ CD31+ hematopoietic progenitors. Bottom row shows gating for mature CD43+ CD45+ hematopoietic progenitors.
- To differentiate iPS cells (or ES cells) into hematopoietic progenitors, prewarm required amounts of collagenase IV solution (1 mg/ml, 37 °C, see Recipes) and KO-DMEM (room temperature, RT) before starting the experiment. Prepare induction medium d0 (see Recipes) and prewarm at RT.
- The differentiation of iPS cell (or ES cell) derived hematopoietic progenitors into DC subsets is summarized in Figure 4.
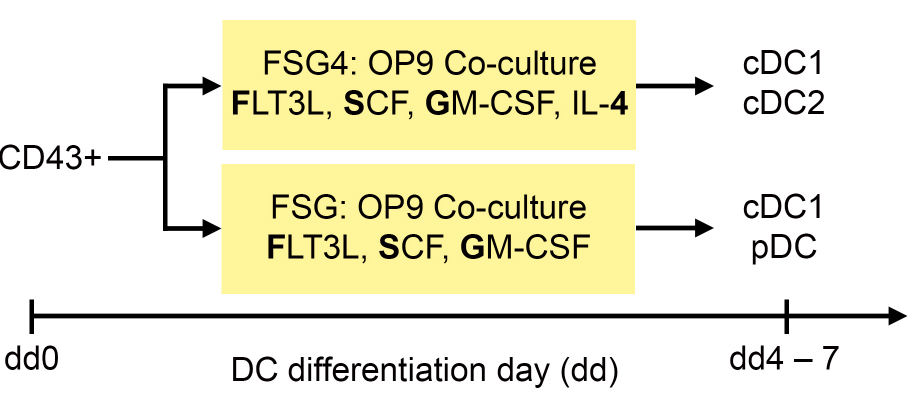
Figure 4. Differentiation protocol of iPS cell (or ES cell) derived hematopoietic progenitors into DC subsets. CD43+ hematopoietic progenitors are co-cultured on OP9 stromal cells with FSG4 and FSG cytokines to generate cDC1 and cDC2 (FSG4), or cDC1 and pDC (FSG), respectively.- To differentiate iPS cell (or ES cell) derived hematopoietic progenitors into DC, seed irradiated OP9 stromal cells (30 Gray) resuspended in OP9 culture medium (see Recipes) in a density of 1.2 x 104 cells/cm2 onto 0.1% gelatin coated TCP dishes one day prior to DC differentiation. We prefer using 6-well plates because later during DC differentiation we can use the same cells for different treatments and experiments.
- Collect hematopoietic progenitors from step A15 by gentle pipetting and pass cell suspension through a 40 µm cell strainer in order to remove remaining EB. Wash plates twice with 1x PBS (prewarmed to RT) and pass this suspension through the same cell strainer.
- Centrifuge flow-through at 416 x g for 4 min. Aspirate supernatant and wash pellet in defined volume of 1x PBS (prewarmed to RT, e.g., 5 ml). Count cells (e.g., with a Neubauer chamber and trypan blue as live/dead cell marker).
- Centrifuge cell suspension at 416 x g for 4 min. Aspirate supernatant and resuspend cells in DC differentiation basal medium (prewarmed to RT, see Recipes) in a density of 1 x 106-1.5 x 106 cells/ml. Split cell suspension in half in two separate Falcon tubes labeled FSG4 and FSG, respectively.
- Add 100 ng/ml FLT3L, 20 ng/ml SCF, 20 ng/ml GM-CSF and 20 ng/ml IL-4 final concentrations in one Falcon tube (FSG4). Add 100 ng/ml FLT3L, 20 ng/ml SCF and 10 ng/ml GM-CSF final concentrations in the other Falcon tube (FSG).
- Aspirate OP9 culture medium from OP9 stromal cells and distribute FSG4 and FSG cell suspension onto the OP9 stromal cells. For example, 6 ml of FSG4 and FSG suspension can be distributed in 3 wells of a 6-well plate (2 ml per well), respectively.
- Culture cells at 37 °C and 5% CO2.
- Perform a partial medium change every second day. Prepare fresh medium: take half of the volume of basal DC differentiation medium as in step B4 but add double the amount of cytokines. Carefully tilt the plate and slowly aspirate the supernatant. Cells are loosely adherent to the feeder layer and those in suspension will settle down to the bottom if the medium is removed slowly. Avoid disturbing the cells e.g., by rinsing them off the feeder layer. Remove approx. 50% of the old medium (e.g., 1 ml per 6-well). Add 50% of freshly prepared medium (see above, prewarmed to RT). Thereby cytokine concentrations in the total volume will be the same as given in step B5.
- DC have acquired DC morphology by DC differentiation day (dd) 4 (Figure 5) and can be used for analyses between dd4-dd7. To collect DC, carefully rinse them off the feeder layer. Wash plates twice with 1x PBS (prewarmed to RT). Avoid detaching OP9 stromal cells. Collect cells in a 50 ml Falcon tube on ice and proceed to required analyses and experiments, respectively.
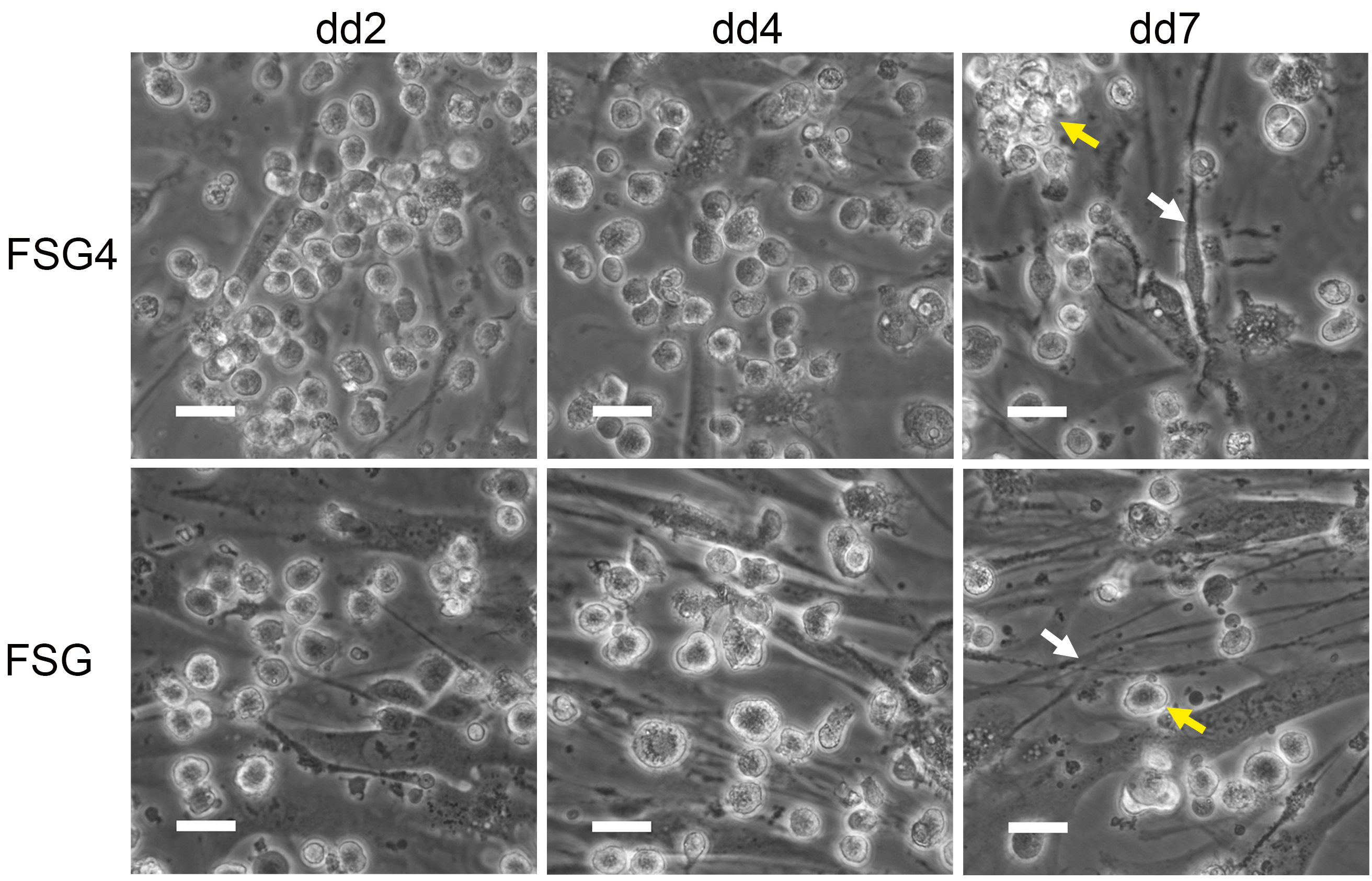
Figure 5. iPS derived DC acquire DC morphology during differentiation. Representative phase contrast images of iPS cell derived DC (yellow arrow) on irradiated OP9 stromal cells (white arrow) during differentiation. Scale bars = 25 µm.
- To differentiate iPS cell (or ES cell) derived hematopoietic progenitors into DC, seed irradiated OP9 stromal cells (30 Gray) resuspended in OP9 culture medium (see Recipes) in a density of 1.2 x 104 cells/cm2 onto 0.1% gelatin coated TCP dishes one day prior to DC differentiation. We prefer using 6-well plates because later during DC differentiation we can use the same cells for different treatments and experiments.
Data analysis
iPS cell (or ES cell) derived DC subsets can be analyzed as described in our recent publication (Sontag et al., 2017).
Notes
Instead of hyper-IL-6 we also tested recombinant human IL-6 (Miltenyi Biotec, catalog number: 130-095-352) in a final concentration of 100 ng/ml and obtained similar results.
Recipes
- Collagenase IV solution (1 mg/ml)
1,000 mg collagenase IV
1,000 ml KO-DMEM
Note: Sterile filter with 0.2 µm top bottle filter. Aliquot (e.g., 50 ml) and store at -20 °C. - 0.1% gelatin solution
0.5 g gelatin
500 ml distilled water
Note: Autoclave at 121 °C for 30 min. Gelatin will solubilize during autoclaving. Store at RT. For coating, cover surface of tissue culture dish with gelatin solution and incubate at 37 °C for at least 15 min. Aspirate gelatin solution prior to plating cells. Use immediately without additional washing or drying. - 1-Thioglycerol stock solution (100 mM)
87 µl 1-thioglycerol
10 ml distilled and sterile water
Note: 1-Thioglycerol has a high viscosity. Pipette slowly in order to dispense 1-thioglycerol accurately. Aliquot (e.g., 1 ml) and store at -20 °C. - L-ascorbic acid stock solution (50 mg/ml)
100 mg L-ascorbic acid
2 ml distilled and sterile water
Note: Aliquot (e.g., 250 µl) and store at -20 °C. Protect from light. - 0.1% BSA solution
0.1 g BSA
100 ml 1x PBS
Note: Store at 4 °C. - BMP4 stock solution (25 µg/ml)
10 µg BMP4
400 µl 0.1% BSA solution - bFGF stock solution (100 µg/ml)
50 µg bFGF
500 µl 0.1% BSA solution - FLT3L stock solution (25 µg/ml)
100 µg FLT3L
4 ml 0.1% BSA solution - GM-CSF stock solution (100 µg/ml)
20 µg GM-CSF
200 µl 0.1% BSA solution - IGF1 stock solution (40 µg/ml)
100 µg IGF1
2.5 ml 0.1% BSA solution - IL-3 stock solution (150 µg/ml)
25 µg IL-3
167 µl 0.1% BSA solution - IL-4 stock solution (20 µg/ml)
10 µg IL-4
500 µl 0.1% BSA solution - SCF stock solution (100 µg/ml)
10 µg SCF
100 µl 0.1% BSA solution - TPO stock solution (20 µg/ml)
100 µg TPO
5 ml 0.1% BSA solution - VEGF stock solution (100 µg/ml)
10 µg VEGF
100 µl 0.1% BSA solutionNote: All cytokines (BMP4, bFGF, FLT3L, GM-CSF, IGF1, IL-3, IL-4, SCF, TPO and VEGF) are stored in aliquots (e.g., 10-50 µl) at -20 °C in a standard non-defrosting freezer. After thawing, we keep them at 4 °C and use them for four weeks.
- Hematopoietic progenitor basal differentiation medium
237 ml 1x StemPro-34 SFM
6.5 ml StemPro-34 supplements (provided with 1x StemPro-34 SFM)
2.5 ml penicillin-streptomycin (final concentration 100 U/ml penicillin, 100 µg/ml streptomycin)
2.5 ml L-glutamine (final concentration 2 mM)
1.0 ml 1-thioglycerol stock solution (final concentration 0.4 mM)
250 µl L-ascorbic acid stock solution (final concentration 50 µg/ml) - Induction medium d0
20 ml hematopoietic progenitor basal differentiation medium
6.7 µl BMP4 stock solution (final concentration 8 ng/ml) - Induction medium d1
20 ml hematopoietic progenitor basal differentiation medium
6.7 µl BMP4 stock solution (final concentration 8 ng/ml)
4.0 µl bFGF stock solution (final concentration 10 ng/ml, see Procedure step A9) - Induction medium d2
20 ml hematopoietic progenitor basal differentiation medium
6.7 µl BMP4 stock solution (final concentration 8 ng/ml)
2.0 µl bFGF stock solution (final concentration 10 ng/ml)
2.0 µl VEGF stock solution (final concentration 10 ng/ml)
20 µl hyper-IL-6 (final concentration 10 ng/ml)
12.5 µl IGF1 stock solution (final concentration 25 ng/ml)
20 µl SCF stock solution (final concentration 100 ng/ml) - Induction medium d4
20 ml hematopoietic progenitor basal differentiation medium
2.0 µl bFGF stock solution (final concentration 10 ng/ml)
2.0 µl VEGF stock solution (final concentration 10 ng/ml)
20 µl hyper-IL-6 (final concentration 10 ng/ml)
12.5 µl IGF1 stock solution (final concentration 25 ng/ml)
20 µl SCF stock solution (final concentration 100 ng/ml)
8.0 µl FLT3L stock solution (final concentration 10 ng/ml)
20 µl TPO stock solution (final concentration 20 ng/ml)
4.0 µl IL-3 stock solution (final concentration 30 ng/ml) - Induction medium d6
20 ml hematopoietic progenitor basal differentiation medium
20 µl SCF stock solution (final concentration 100 ng/ml)
8.0 µl FLT3L stock solution (final concentration 10 ng/ml)
20 µl TPO stock solution (final concentration 20 ng/ml)
4.0 µl IL-3 stock solution (final concentration 30 ng/ml) - OP9 culture medium
195 ml α-MEM
50 ml FBS (final concentration 20%)
2.5 ml penicillin-streptomycin (final concentration 100 U/ml penicillin, 100 µg/ml streptomycin)
2.5 ml L-glutamine (final concentration 2 mM) - DC differentiation basal medium
220 ml RPMI 1640
25 ml FBS (final concentration 10%)
2.5 ml penicillin-streptomycin (final concentration 100 U/ml penicillin, 100 µg/ml streptomycin)
2.5 ml L-glutamine (final concentration 2 mM)
500 µl β-mercaptoethanol (final concentration 0.1 mM) - DC differentiation FSG4 medium
20 ml DC differentiation basal medium
80 µl FLT3L stock concentration (final concentration 100 ng/ml)
40 µl SCF stock concentration (final concentration 20 ng/ml)
4.0 µl GM-CSF stock solution (final concentration 20 ng/ml)
20 µl IL-4 stock concentration (final concentration 20 ng/ml) - DC differentiation FSG medium
20 ml DC differentiation basal medium
80 µl FLT3L stock concentration (final concentration 100 ng/ml)
40 µl SCF stock concentration (final concentration 20 ng/ml)
2.0 µl GM-CSF stock solution (final concentration 20 ng/ml)
Acknowledgments
This work was supported in part by the Ministry for Innovation, Science and Research of German Federal State of North Rhine-Westphalia, Duesseldorf, Germany (S. S. and M. Z.) and by a research grant of the Interdisciplinary Center for Clinical Research (IZKF) Aachen, Germany (M. Z.). We thank Marion Kennedy and Gordon Keller for their advice on hematopoietic progenitor differentiation.
References
- Bellin, M., Marchetto, M. C., Gage, F. H. and Mummery, C. L. (2012). Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol 13(11): 713-726.
- Belz, G. T. and Nutt, S. L. (2012). Transcriptional programming of the dendritic cell network. Nat Rev Immunol 12(2): 101-113.
- Choi, K. D., Vodyanik, M. and Slukvin, II (2011). Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat Protoc 6(3): 296-313.
- Fischer, M., Goldschmitt, J., Peschel, C., Brakenhoff, J. P., Kallen, K. J., Wollmer, A., Grotzinger, J. and Rose-John, S. (1997). A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol 15(2): 142-145.
- Gilliet, M., Boonstra, A., Paturel, C., Antonenko, S., Xu, X. L., Trinchieri, G., O'Garra, A. and Liu, Y. J. (2002). The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med 195(7): 953-958.
- Grskovic, M., Javaherian, A., Strulovici, B. and Daley, G. Q. (2011). Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov 10(12): 915-929.
- Joffre, O. P., Segura, E., Savina, A. and Amigorena, S. (2012). Cross-presentation by dendritic cells. Nat Rev Immunol 12(8): 557-569.
- Jongbloed, S. L., Kassianos, A. J., McDonald, K. J., Clark, G. J., Ju, X., Angel, C. E., Chen, C. J., Dunbar, P. R., Wadley, R. B., Jeet, V., Vulink, A. J., Hart, D. N. and Radford, K. J. (2010). Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med 207(6): 1247-1260.
- Kennedy, M., Awong, G., Sturgeon, C. M., Ditadi, A., LaMotte-Mohs, R., Zuniga-Pflucker, J. C. and Keller, G. (2012). T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2(6): 1722-1735.
- Lee, J., Breton, G., Oliveira, T. Y., Zhou, Y. J., Aljoufi, A., Puhr, S., Cameron, M. J., Sekaly, R. P., Nussenzweig, M. C. and Liu, K. (2015). Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med 212(3): 385-399.
- Li, Y., Liu, M. and Yang, S. T. (2014). Dendritic cells derived from pluripotent stem cells: Potential of large scale production. World J Stem Cells 6(1): 1-10.
- Lundberg, K., Albrekt, A. S., Nelissen, I., Santegoets, S., de Gruijl, T. D., Gibbs, S. and Lindstedt, M. (2013). Transcriptional profiling of human dendritic cell populations and models--unique profiles of in vitro dendritic cells and implications on functionality and applicability. PLoS One 8(1): e52875.
- Merad, M., Sathe, P., Helft, J., Miller, J. and Mortha, A. (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 31: 563-604.
- Poulin, L. F., Salio, M., Griessinger, E., Anjos-Afonso, F., Craciun, L., Chen, J. L., Keller, A. M., Joffre, O., Zelenay, S., Nye, E., Le Moine, A., Faure, F., Donckier, V., Sancho, D., Cerundolo, V., Bonnet, D. and Reis e Sousa, C. (2010). Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells. J Exp Med 207(6): 1261-1271.
- Robinton, D. A. and Daley, G. Q. (2012). The promise of induced pluripotent stem cells in research and therapy. Nature 481: 295-305.
- Rossi, R., Hale, C., Goulding, D., Andrews, R., Abdellah, Z., Fairchild, P. J. and Dougan, G. (2012). Interaction of Salmonella typhimurium with dendritic cells derived from pluripotent embryonic stem cells. PLoS One 7(12): e52232.
- Schlitzer, A. and Ginhoux, F. (2014). Organization of the mouse and human DC network. Curr Opin Immunol 26: 90-99.
- Schmid, M. A., Kingston, D., Boddupalli, S. and Manz, M. G. (2010). Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev 234(1): 32-44.
- Senju, S., Haruta, M., Matsumura, K., Matsunaga, Y., Fukushima, S., Ikeda, T., Takamatsu, K., Irie, A. and Nishimura, Y. (2011). Generation of dendritic cells and macrophages from human induced pluripotent stem cells aiming at cell therapy. Gene Ther 18(9): 874-883.
- Senju, S., Suemori, H., Zembutsu, H., Uemura, Y., Hirata, S., Fukuma, D., Matsuyoshi, H., Shimomura, M., Haruta, M., Fukushima, S., Matsunaga, Y., Katagiri, T., Nakamura, Y., Furuya, M., Nakatsuji, N. and Nishimura, Y. (2007). Genetically manipulated human embryonic stem cell-derived dendritic cells with immune regulatory function. Stem Cells 25(11): 2720-2729.
- Silk, K. M., Silk, J. D., Ichiryu, N., Davies, T. J., Nolan, K. F., Leishman, A. J., Carpenter, L., Watt, S. M., Cerundolo, V. and Fairchild, P. J. (2012). Cross-presentation of tumour antigens by human induced pluripotent stem cell-derived CD141+XCR1+ dendritic cells. Gene Ther 19(10): 1035-1040.
- Sontag, S., Forster, M., Qin, J., Wanek, P., Mitzka, S., Schuler, H. M., Koschmieder, S., Rose-John, S., Sere, K. and Zenke, M. (2017). Modelling IRF8 deficient human hematopoiesis and dendritic cell development with engineered iPS cells. Stem Cells 35(4): 898-908.
- Swiecki, M. and Colonna, M. (2015). The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15(8): 471-485.
- Takahashi, K. and Yamanaka, S. (2016). A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 17(3): 183-193.
- Tseng, S. Y., Nishimoto, K. P., Silk, K. M., Majumdar, A. S., Dawes, G. N., Waldmann, H., Fairchild, P. J., Lebkowski, J. S. and Reddy, A. (2009). Generation of immunogenic dendritic cells from human embryonic stem cells without serum and feeder cells. Regen Med 4(4): 513-526.
- Villadangos, J. A. and Shortman, K. (2010). Found in translation: the human equivalent of mouse CD8+ dendritic cells. J Exp Med 207:1131-1134.
- Yanagimachi, M. D., Niwa, A., Tanaka, T., Honda-Ozaki, F., Nishimoto, S., Murata, Y., Yasumi, T., Ito, J., Tomida, S., Oshima, K., Asaka, I., Goto, H., Heike, T., Nakahata, T. and Saito, M. K. (2013). Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS One 8(4): e59243.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sontag, S., Förster, M., Seré, K. and Zenke, M. (2017). Differentiation of Human Induced Pluripotent Stem Cells (iPS Cells) and Embryonic Stem Cells (ES Cells) into Dendritic Cell (DC) Subsets. Bio-protocol 7(15): e2419. DOI: 10.21769/BioProtoc.2419.
Category
Stem Cell > Pluripotent stem cell > Cell induction
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











