- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Multiplex Gene Editing via CRISPR/Cas9 System in Sheep
Published: Vol 7, Iss 13, Jul 5, 2017 DOI: 10.21769/BioProtoc.2385 Views: 11639
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Multiplex Genome Editing of Human Pluripotent Stem Cells Using Cpf1
Haiting Ma
Nov 20, 2024 2650 Views

Reprogramming White Fat Cells for Adipose Manipulation Transplantation (AMT) Therapy
Kelly An [...] Nadav Ahituv
Aug 5, 2025 2240 Views

Generation of Insulin-Producing Alpha TC1-6 Cells Using EpiCRISPR System for Targeted DNA Methylation
Marija B. Đorđević [...] Melita S. Vidaković
Oct 20, 2025 1232 Views
Abstract
Sheep is a major large animal model for studying development and disease in biomedical research. We utilized CRISPR/Cas9 system successfully to modify multiple genes in sheep. Here we provide a detailed protocol for one-cell-stage embryo manipulation by co-injecting Cas9 mRNA and RNA guides targeting three genes (MSTN, ASIP, and BCO2) to create genetic-modified sheep. Procedure described sgRNA design, construction of gRNA-Cas9 plasmid, efficient detection in fibroblast, embryos and sheep, and some manipulative technologies. Our findings suggested that the CRISPR/Cas9 method can be exploited as a powerful tool for livestock improvement by targeting multiple genes that are in charge of economically significant traits simultaneously.
Keywords: Cas9Background
Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have been used to modify many cell lines and organisms in the past. The recent CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated) technology development provides an efficient tool for genome modifying of targeting loci. This system has an advantage of modifying multiple loci at the same time. Sheep with specific gene modifications may contribute to breeding. The results demonstrate the first detailed evidence of large animal modification in sheep.
Materials and Reagents
- Pipette tips
- Centrifuge tubes
- 6-well culture plate
- HiBindTM DNA Mini Column
- Sheep (Healthy ewes [3-5 years old] were selected about 160)
- PGL3-U6-gRNA vector (Addgene, catalog number: 51133 )
- pUC57-T7-gRNA vector (Addgene, catalog number: 51132 )
- E. coli DH5α competent cells
- Cas9 mRNA in vitro transcription vector (Addgene, catalog number: 44758 )
- T vector (Takara Bio, catalog number: D103A )
- Bsal enzyme(New England Biolabs, catalog number: R3535 )
- DNA Gel Extraction Kit (Corning, catalog number: D205-04 )
- T4 ligase (New England Biolabs)
- Ampicillin
- EndoFree Plasmid Maxi Kit (QIAGEN, catalog number: 12362 )
- Primers
- M13R (-47) primer
5’-CGCCAGGGTTTTCCCAGTCACGAC-3’
- Primers for amplifying the selected potential off-target loci, designed by Primer5 software.
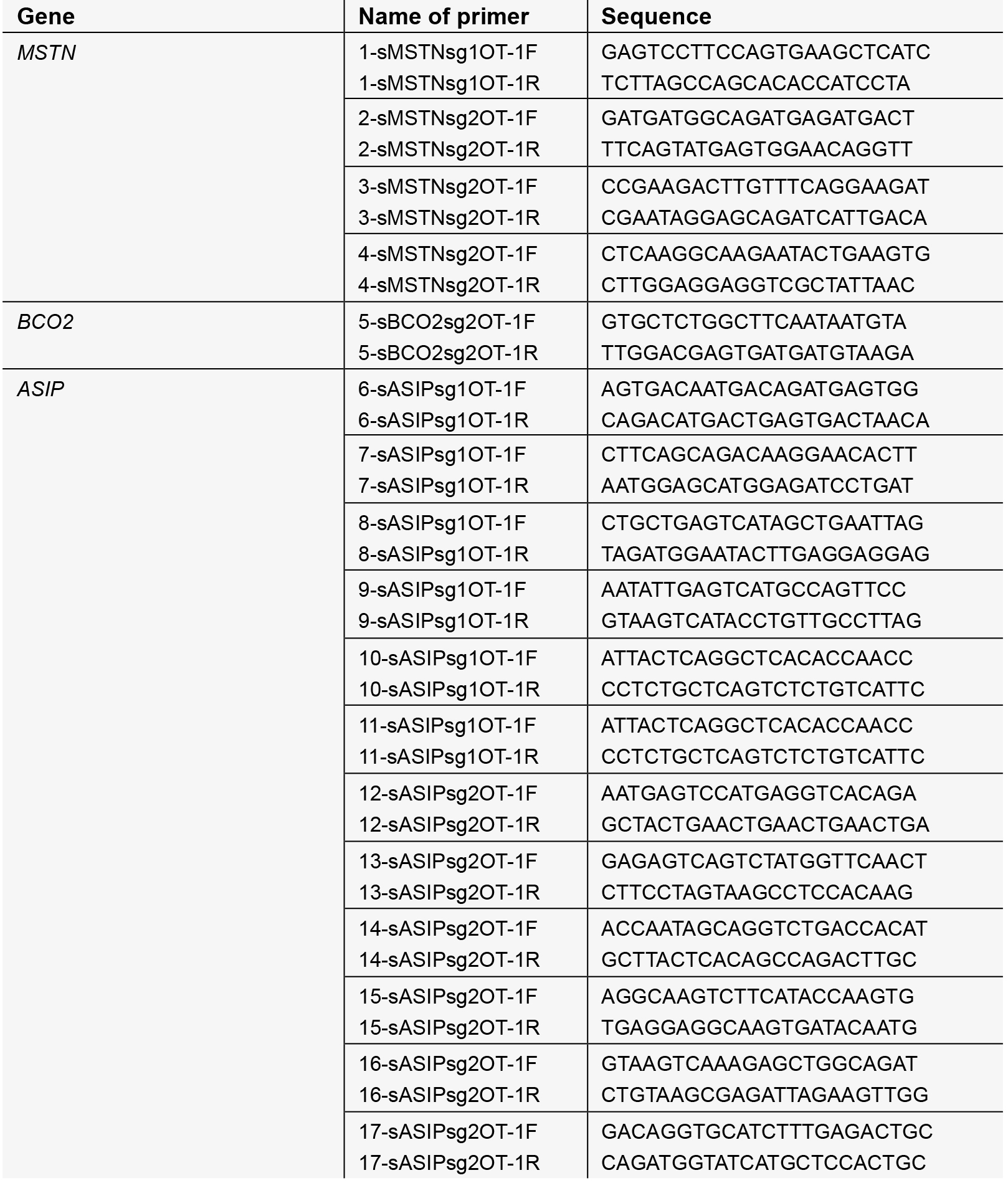
- Ethanol
- Ice acetic acid (Guangzhou Chemical Reagent Factory, catalog number: CB39-GR-0.5L )
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM)
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM)
- Lipofectamine3000 reagent (Thermo Fisher Scientific, InvitrogenTM)
- Opti-MEM medium (Thermo Fisher Scientific, GibcoTM)
- Blasticidine S hydrochloride
- Universal Genomic DNA Kit (CWBIO, catalog number: CW2298M )
- KOD-Plus-Neo
- PCR cleanup kit (AppliChem, catalog number: A7089,1000 )
- T7EI
- Agarose
- TCM199 medium (Thermo Fisher Scientific, GibcoTM)
- Quinn’s Advantage Cleavage Medium and Blastocyst Medium (Sage Biopharma)
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific)
- REPLI-g Single Cell Kit (QIAGEN, catalog number: 150343 )
- D2000 DNA marker
- Tris
- Ethylenediaminetetraacetate acid (EDTA)
- Agar powder
- Tryptone
- Yeast extract
- Sodium chloride (NaCl)
- 50x TAE (see Recipes)
- LB medium (see Recipes)
Equipment
- Pipettes
- 37 °C water bath
- Thermal cycler (Bio-Rad thermocycler)
- DNA electrophoresis apparatus
- Microcentrifuge
- Eppendorf FemtoJect system
- Olympus micromanipulation system ON3 (Olympus, model: ON3 SERIES )
- Microinjection device
- Biopsy forceps (Olympus, model: FB-11K-1 )
Software
- Primer5 software
Procedure
- sgRNA design
- Select specific target sites in sheep genome and then design sgRNA-oligo sequence.
- After design of sgRNA-oligo sequence, appropriate adaptor sequences should be added for cloning. The upstream primer of both pUC57-T7-gRNA vector and PGL3-U6-gRNA should include 5’-TAGG-3’ adaptor sequence, while the downstream primer for both should contain an adaptor with the sequence of 5’-AAAC-3’.
- Select specific target sites in sheep genome and then design sgRNA-oligo sequence.
- Construct sgRNA-Cas9 plasmid
- Digest the PGL3-U6-gRNA vector and pUC57-T7-gRNA vector by BsaI.

- Incubate at 37 °C for 3-4 h.
- Purify the digested vector using DNA Gel Extraction Kit.
- Double-strand sequences and prepare double-strand oligo DNA reaction system:
MSTN:
sgRNA1:
5’ CCAACTGTGGATATATCTGAGAC 3’
5’ GTCTCAGATATATCCACAGTTGG 3’
sgRNA2:
5’ GGATTTTGAAGCTTTTGGATGG 3’
5’ CCATCCAAAAGCTTCAAAATCC 3’
ASIP:
sgRNA1:
5’ CCAGAGATGAAAGGAACCTGAAG 3’
5’ CTTCAGGTTCCTTTCATCTCTGG 3’
sgRNA2:
5’ CAATTCTTCCATGAACCTGTTGG 3’
5’ CCAACAGGTTCATGGAAGAATTG 3’
BCO2:
sgRNA1:
5’ CCATGCATTGCACCGCTTCTAAC 3’
5’ GTTAGAAGCGGTGCAATGCATGG 3’
sgRNA2:
5’ GGTCGTCTCAGCTCGAGTCCAGG 3’
5’ CCTGGACTCGAGCTGAGACGACC 3’

- Perform PCR using the following cycling conditions:

- Ligate the diluted double-strand oligo DNA with the PGL3-U6-gRNA vector digested by BsaI:

- Incubate at 16 °C overnight.
- Transform 1 µl of ligation product into 50 µl E. coli DH5α competent cells.
- Pick up one colony and inoculate in a 15 ml tube containing 5 ml LB medium with 50 µg/ml ampicillin (4 colonies for each transformation). Incubate overnight in a 37 °C shaker.
- Extract plasmids from transformed DH5α cells using Endo-Free Plasmid Mini Kit following the manufacturer’s instructions (see Supplement 1).
- Confirm the positive clones containing PGL3-U6-gRNA and pUC57-T7-gRNA by Sanger sequencing using M13R (-47) primer.
- Digest the PGL3-U6-gRNA vector and pUC57-T7-gRNA vector by BsaI.
- Cas9/sgRNA efficacy test in sheep fibroblast
- Sheep fibroblasts were obtained from a 40-day-old fetus and then transported to the laboratory.
- The fibroblasts are cultured for five passages in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Gibco) until 80-90% confluence.
- Cell transfection.
- The transfection procedure is performed using Lipofectamine3000 reagent (Invitrogen) according to the specification (Table 1).
Table 1. The specification about transfection using Lipofectamine3000 Reagent (Invitrogen)
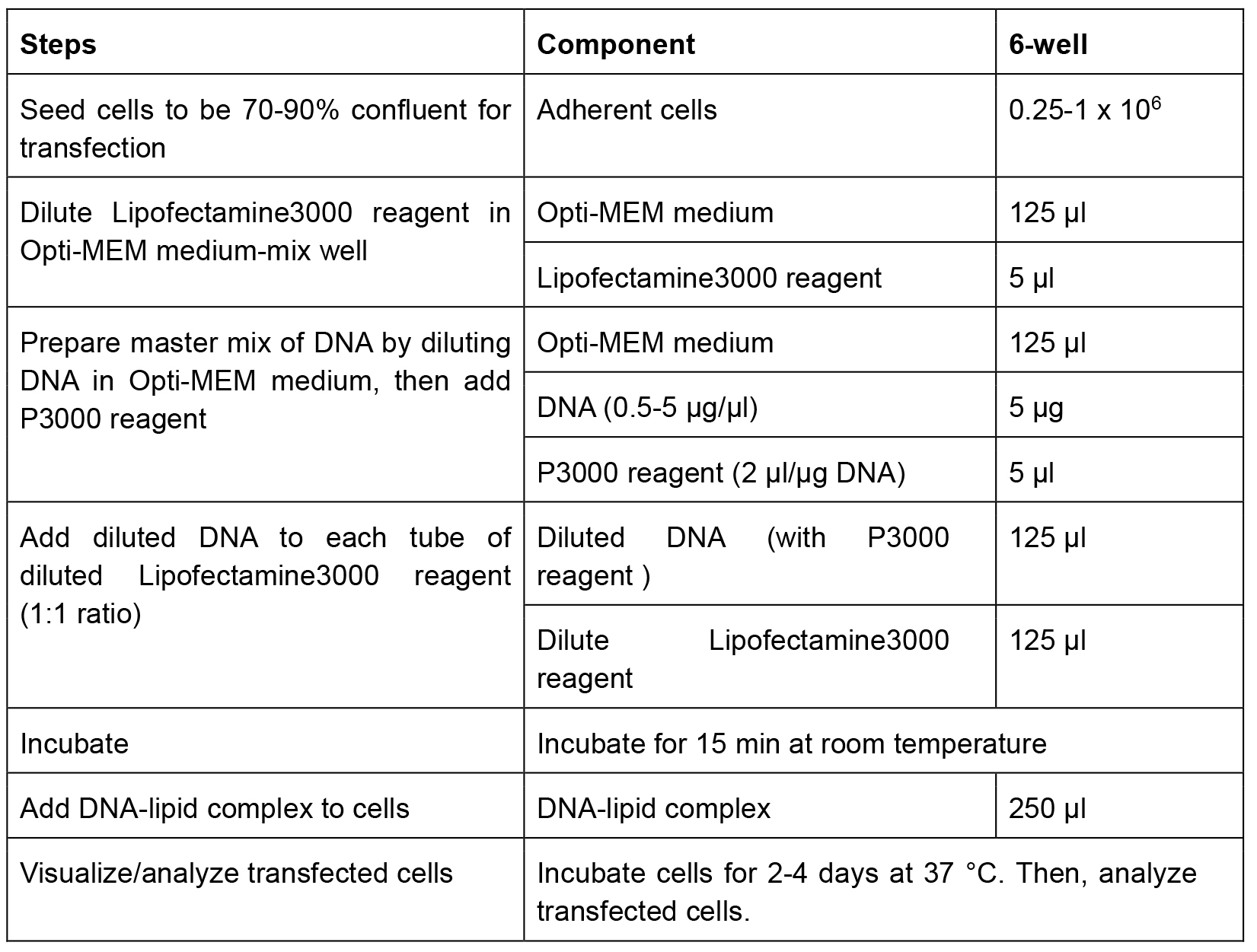
- Fibroblasts are separately transfected as followed types:

- The transfection procedure is performed using Lipofectamine3000 reagent (Invitrogen) according to the specification (Table 1).
- Dilute by adding 10 mg/ml of blasticidin S hydrochloride into the medium (1:1,000 dilution) 24 h after transfection.
- Then incubate for 24 h at 37 °C.
- Genomic DNA is extracted from fibroblasts at 72 h after transfection using a Universal Genomic DNA Kit.
- Amplify targeted fragments using KOD polymerase, then purify with a PCR cleanup kit.
- Primers used for amplifying Cas9/sgRNA targeted MSTN, ASIP and BCO2 fragment.
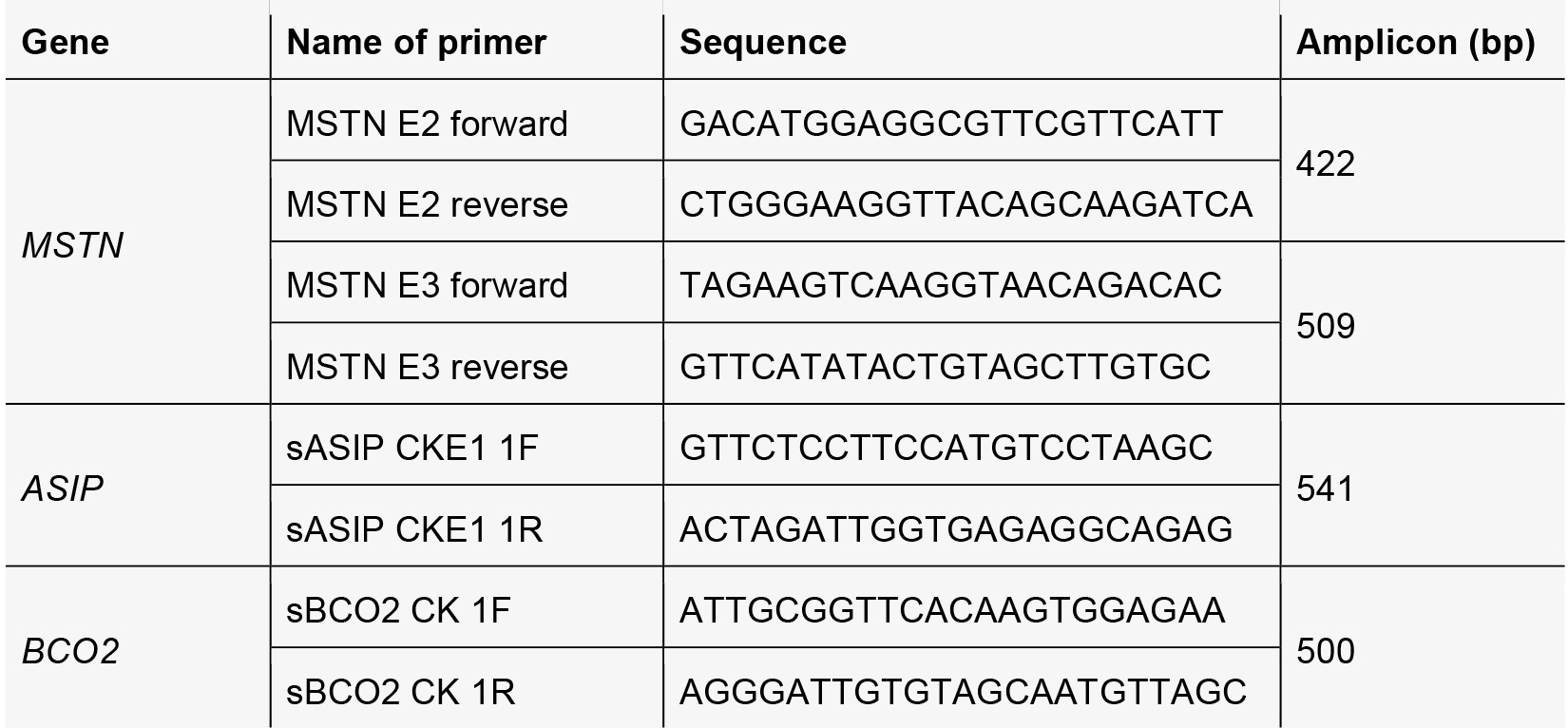
- Prepare the reaction mixtures as follows:

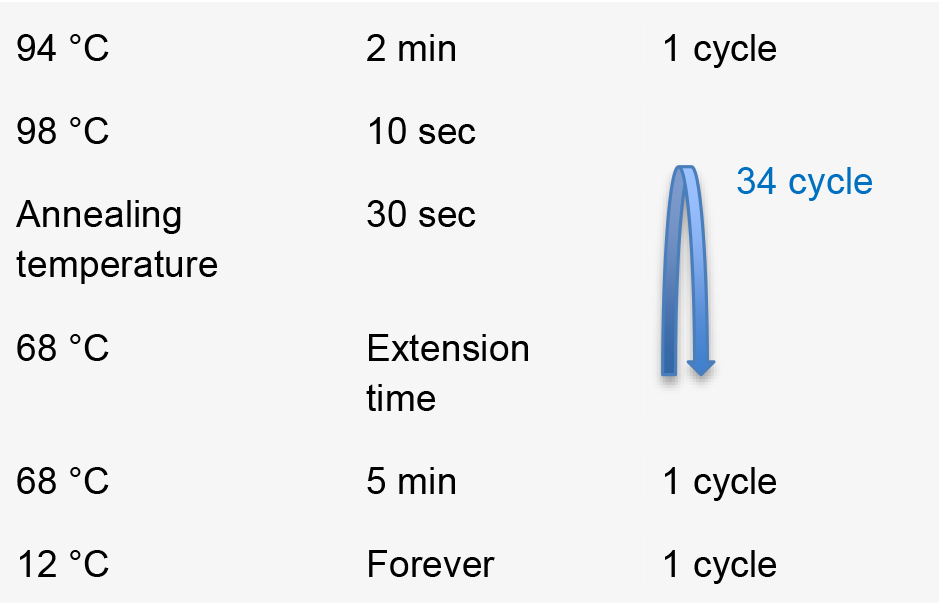
- Perform PCR using the following conditions [annealing temperature (T) and extension time (L)].
- Primers used for amplifying Cas9/sgRNA targeted MSTN, ASIP and BCO2 fragment.
- The purified PCR products are denatured and re-annealed in NEBuffer 2 (NEB) using a Bio-Rad thermocycler.
- The PCR products are digested with T7EI for 30 min at 37 °C and separated by 2.5% agarose gel electrophoresis.
- PCR products of mutations detected by T7EI cleavage assay are sub-cloned into T vector.
- For each sample, the colonies are randomly picked and sequenced using an M13F (-47) to confirm the positive colony.
- Sheep fibroblasts were obtained from a 40-day-old fetus and then transported to the laboratory.
- Off-target detection in sheep fibroblasts
- Predict the potential off-target sites using SeqMap.
- Extract sheep fibroblasts DNA for PCR amplification using specific primers (see Materials and Reagents).
- Mix the reaction components according to the follows:
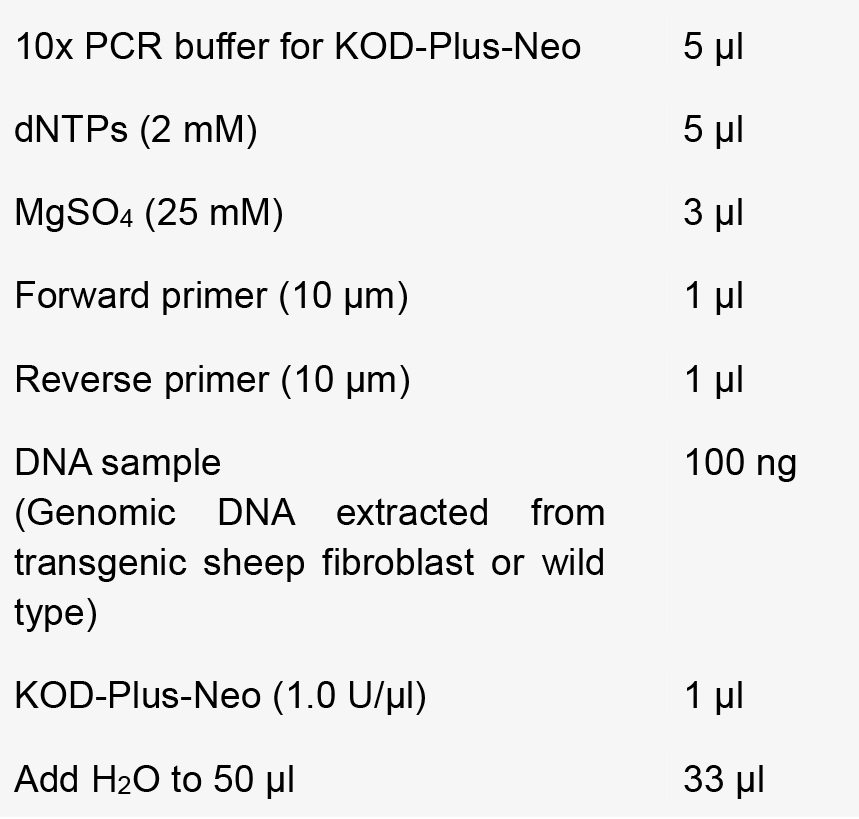
- Perform PCR using the following conditions [annealing temperature (T) and extension time (L)].

- Mix the reaction components according to the follows:
- The PCR products are then subjected to a T7EI cleavage assay as described above and results below (Figure 1).
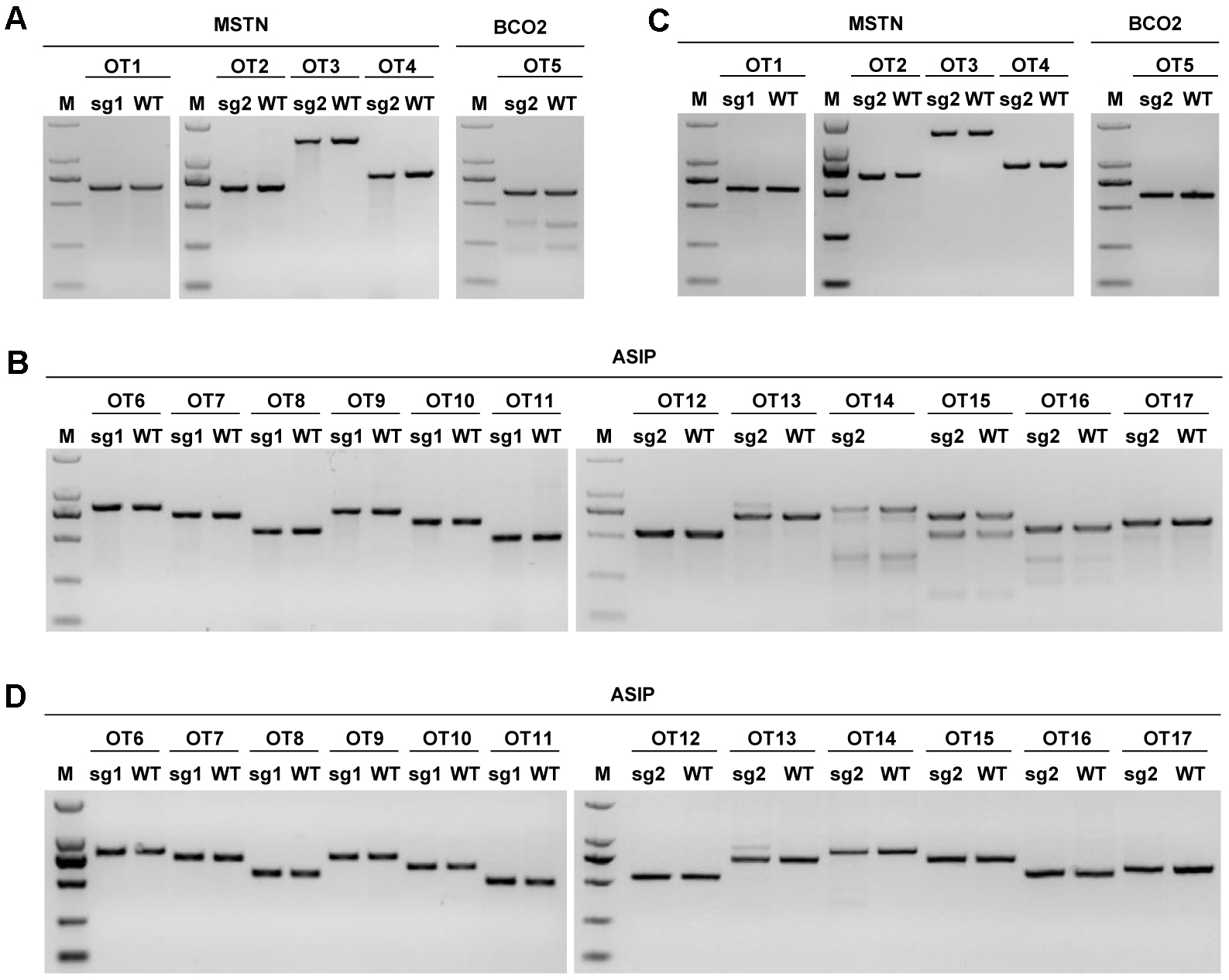
Figure 1. Detection of the sgRNA:Cas9-mediated off-target cleavage of MSTN, ASIP, and BCO2 by using the T7EI cleavage assay. The PCR products of the potential off-target sites in MSTN, ASIP, and BCO2 sgRNA:Cas9 loci from sheep fibroblasts (A and B). 17 predicted off-target sites that were most homologous to MSTN, ASIP, and BCO2 sgRNAs were named OT1 to OT17. C and D. Detection of the sgRNA:Cas9-mediated off-target cleavage of MSTN, ASIP, and BCO2 by using the T7EI cleavage assay. More messages see Wang et al. (2016).
- Predict the potential off-target sites using SeqMap.
- Zygote collection
Note: Healthy ewes (3-5 years old) were selected to be desired donors for zygote collection which include regular estrus cycles.- Zygote collection and treatment of donors are conducted as earlier described in goats (Wang et al., 2015).
- Collect one cell stage zygotes (around 10-16 h after the last insemination) using a surgical way and immediately transfer the collected zygotes into a TCM199 medium.
- Zygote collection and treatment of donors are conducted as earlier described in goats (Wang et al., 2015).
- Efficient detection of sheep zygotes
- Microinject the sgRNA & Cas9 mRNA into sheep zygotes using an Olympus micromanipulation system ON3 in a manipulation medium TCM199 on the heated platform.
Note: Mix Cas9 mRNA (20 ng/µl) and sgRNAs (5 ng/µl for each sgRNA) targeting MSTN, ASIP and BCO2.
- Inject the sgRNA & Cas9 mRNA into the cytoplasm of fertilized oocytes using an Eppendorf FemtoJect system with the following set-up.

- The embryos are cultured for 168 h at 38.5 °C in Quinn’s Advantage Cleavage Medium and Blastocyst Medium (Sage Biopharma).
- Blastocyst embryos are used for efficient detection preserved in PBS.
- Genomes of embryos are amplified using REPLI-g Single Cell Kit.
- Targeted fragments are amplified as described above including primers, reactions and procedures.
- Purification of PCR products, T7EI cleavage assay, and Sanger sequencing are performed according to Wang et al. (2015).
- Microinject the sgRNA & Cas9 mRNA into sheep zygotes using an Olympus micromanipulation system ON3 in a manipulation medium TCM199 on the heated platform.
- Production of gene-modified sheep
Note: Gene-modified sheep and control animals were fed the same diet, and managed at the same conditions throughout the experiment.- Select the surrogate ewes with their natural estrus cycles as gist.
- Zygote collection and treatment of donors are conducted as described above.
- The zygotes are cultured for 24 h at 37 °C using Quinn’s Advantage Cleavage Medium (Sage Biopharma).
- Then culture the zygotes in Quinn’s Advantage Blastocyst Medium (Sage Biopharma) at 37 °C, 5% CO2 and saturated humidity.
- About 3-4 divisive embryos are transplanted into the ampullary-isthmic junction of the oviduct of the recipient ewes.
- Pregnancy normally takes about 150 days in Tan sheep, and is determined based on the observation of estrus behaviors in every ovulation cycle.
- Select the surrogate ewes with their natural estrus cycles as gist.
- Efficiency detection in Genetic-modified sheep
- Jugular venous blood samples and tissues are collected from randomly selected genetic-modified sheep (n = 10) and randomly selected control animals (n = 10) at 240 days of age for blood and tissues chemistry analyses.
- Genomic DNA is extracted from blood and tissues using corresponding assay from the University Genomic DNA Kit.
- Targeted fragments are amplified as described above in off-target detection in sheep fibroblast (including primers, reactions and procedures) (Procedure D).
- T7EI cleavage assay and sequencing are performed as previously described (step C9).
- The results may confirm the mutation in gene-modified sheep. (Figure 2)
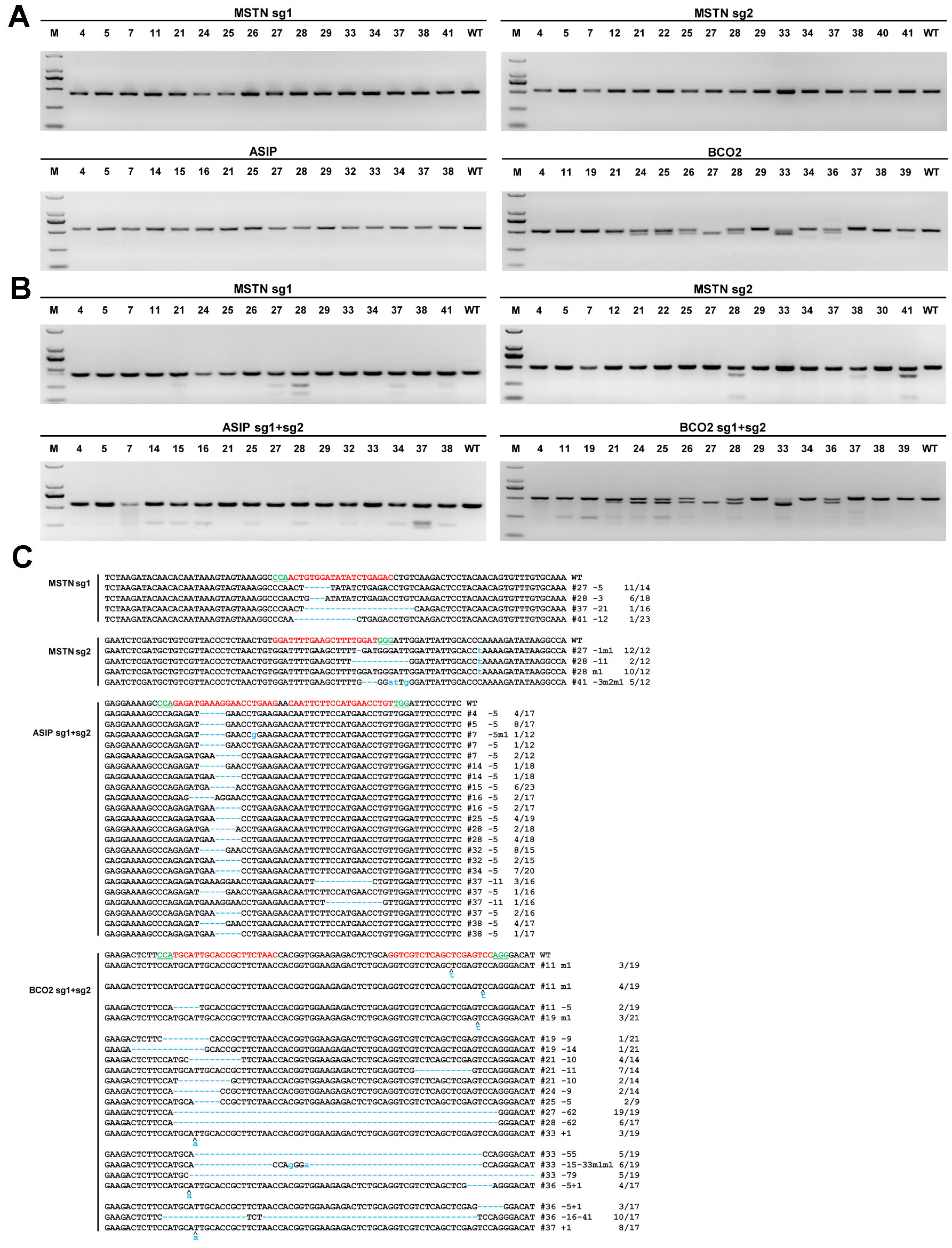
Figure 2. Evaluation of sgRNA:Cas9-mediated genetic modifications in lambs. A. PCR products of the targeted region of the MSTN, ASIP, and BCO2 genes of founder sheep co-microinjected with a mixture of Cas9 mRNA and sgRNAs. D2000 DNA marker was used as a marker reference. B. Detection of sgRNA:Cas9-mediated on-target cleavage of the MSTN, ASIP and BCO2 genes by using a T7EI cleavage assay. All PCR products from (A) were subjected to the T7EI cleavage assay. C. Sequencing results of the modified MSTN, ASIP and BCO2 loci that were detected in the founder animals. Target sequences complementary to sgRNAs of targeted genes are in red text, while the PAM sequences are marked in green. The mutations are marked in blue, dashlines indicates deletions, and lowercase indicates insertions or replacements. Insertions (+), deletions (-), mutations (m) is shown to the right of each allele. The genotypes are shown to the right with the rates of total clones for TA-sequencing. For more information see Wang et al. (2016).
- Jugular venous blood samples and tissues are collected from randomly selected genetic-modified sheep (n = 10) and randomly selected control animals (n = 10) at 240 days of age for blood and tissues chemistry analyses.
- Germline transmission detection
- Germ cells from the testis of the three founders (#28, #33, and #37) were obtained by using biopsy forceps.
- The genomes of germ cells are amplified using REPLI-g Single Cell Kit.
- PCR products with mutations are detected using the T7EI cleavage assay, followed by Sanger sequencing.
- Germ cells from the testis of the three founders (#28, #33, and #37) were obtained by using biopsy forceps.
Recipes
- 50x TAE
Dissolve 242 g of Tris, 37.2 g of EDTA in 700 ml dH2O
Note: Heat solution to 60 °C to assist in dissolution.
Add 57.1 ml of ice acetic acid
Add dH2O to 1,000 ml
Then 200 ml solution was diluted to 1x TAE working solution by adding 10 L dH2O
- LB medium
Dissolve 2 g agar powder, 2 g tryptone, 1 g yeast extract, 1 g NaCl in 200 ml dH2O
Acknowledgments
This work was supported by National Natural Science Foundation of China (31372279, 31402038, and 31171377), the Major Projects for New Varieties of Genetically Modified Organisms of China (2014ZX08008-002), as well as by China Agriculture Research System (CARS-40-13).
References
- Wang, X., Niu, Y., Zhou, J., Yu, H., Kou, Q., Lei, A., Zhao, X., Yan, H., Cai, B., Shen, Q., Zhou, S., Zhu, H., Zhou, G., Niu, W., Hua, J., Jiang, Y., Huang, X., Ma, B. and Chen, Y. (2016). Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci Rep 6: 32271.
- Wang, X., Yu, H., Lei, A., Zhou, J., Zeng, W., Zhu, H., Dong, Z., Niu, Y., Shi, B., Cai, B., Liu, J., Huang, S., Yan, H., Zhao, X., Zhou, G., He, X., Chen, X., Yang, Y., Jiang, Y., Shi, L., Tian, X., Wang, Y., Ma, B., Huang, X., Qu, L. and Chen, Y. (2015). Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep 5: 13878.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Niu, Y., Ding, Y., Wang, X. and Chen, Y. (2017). Multiplex Gene Editing via CRISPR/Cas9 System in Sheep. Bio-protocol 7(13): e2385. DOI: 10.21769/BioProtoc.2385.
Category
Molecular Biology > DNA > Mutagenesis
Molecular Biology > RNA > Transfection
Cell Biology > Cell engineering > CRISPR-cas9
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








