- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single-molecule RNA Fluorescence in situ Hybridization (smFISH) in Caenorhabditis elegans
Published: Vol 7, Iss 12, Jun 20, 2017 DOI: 10.21769/BioProtoc.2357 Views: 18504
Reviewed by: Peichuan ZhangPrashanth SuravajhalaPia Giovannelli

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Adapting the Smart-seq2 Protocol for Robust Single Worm RNA-seq
Lorrayne Serra [...] Ali Mortazavi
Feb 20, 2018 20806 Views

In situ Hybridization (ISH) in Preparasitic and Parasitic Stages of the Plant-parasitic Nematode Meloidogyne spp.
Maëlle Jaouannet [...] Bruno Favery
Mar 20, 2018 9848 Views

Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Ruben Warkentin and Anna Marie Pyle
Oct 20, 2025 1392 Views
Abstract
Single-molecule RNA fluorescence in situ hybridization (smFISH) is a technique to visualize individual RNA molecules using multiple fluorescently-labeled oligonucleotide probes specific to the target RNA (Raj et al., 2008; Lee et al., 2016a). We adapted this technique to visualize RNAs in the C. elegans whole adult worm or its germline, which enabled simultaneous recording of nascent transcripts at active transcription sites and mature mRNAs in the cytoplasm (Lee et al., 2013 and 2016b). Here we describe each step of the smFISH procedure, reagents, and microscope settings optimized for C. elegans extruded gonads.
Keywords: Active transcription siteBackground
smFISH enables direct and precise quantitation of mRNA in vivo. In addition, multiple RNA species can be scored simultaneously in the same cell by multiplexing smFISH probes. There have been previous publications using smFISH in C. elegans, but those studies used wide-field microscopy, which often has lower spatial resolution and requires additional image processing (e.g., image deconvolution) (Ji and van Oudenaarden, 2012). Here we describe an smFISH procedure optimized for the C. elegans germline tissue. Each step is detailed, including use of confocal microscopy to obtain precise measurements. Our protocol minimizes sample-to-sample variability and allows precise quantitation of mRNA and nascent transcripts.
Materials and Reagents
- Gloves
- RNase-free microcentrifuge tubes (1.5 ml) (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: AM12450 )
- Microscope slide (Fisher Scientific, catalog number: 12-544-1 )
- High-precision microscope cover glass (Marienfeld-Superior, catalog number: 0107052 ; can be ordered through Azer Scientific)
Note: Any cover glass compatible with the microscope used for smFISH can be used. - Kimwipes (KCWW, Kimberly-Clark, catalog number: 34155 )
- Aluminum foil
- Scalpel (Feather disposable scalpel #10) (Medex Supply, catalog number: GRF-2975#10 ) or needle (25 G) (BD, catalog number: 305125 )
- RNase-free filtered tips (Mettler-Toledo, Rainin, catalog numbers: 17007957 , 17002927 and 17014361 for 20, 200 and 1,000 µl tips, respectively)
- 0.2 µm syringe filter (EMD Millipore, catalog number: SCGP00525 )
- C. elegans. Some strains were provided by the CGC (http://www.cgc.cbs.umn.edu, which is funded by NIH Office of Research Infrastructure Programs [P40 OD010440])
- smFISH probe(s) conjugated with fluorophores (LGC BioSearch Technologies, https://www.biosearchtech.com/), see below for probe storage and dilution
- RNaseZap® (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9780 )
- ProLong Gold Antifade Reagent Mountant (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36930 )
- Nail polish
- RNase-free 1x PBS (Fisher Scientific, catalog number: BP24384 )
- Tween-20 (Fisher Scientific, catalog number: BP337-100 )
- Levamisole (or Tetramisole) (Sigma-Aldrich, catalog number: L9756-10G ); store at -20 °C
- 37% formaldehyde (AMRESCO, catalog number: 0493-500ML )
- Triton X-100 (Fisher Scientific, catalog number: BP151-100 )
- Ethanol (Acros Organics, catalog number: 615090010 )
- Tris base (Fisher Scientific, catalog number: BP152-5 )
- 0.5 M EDTA, pH 8.0 (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9260G )
- SSC (20x) (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9763 )
- Formamide, deionized (EMD Millipore, catalog number: 4610-100ML ); store at 4 °C; warm to room temperature prior to opening
- DEPC water, sterile (EMD Millipore, catalog number: 9610-1L )
- 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D1306 )
- Dextran sulfate (VWR, catalog number: 97061-196 )
- Glucose, nuclease free (Fisher Scientific, catalog number: D16-500 )
- Tris, pH 8.0, nuclease free (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9855G )
- Glucose oxidase (MP Biomedicals, catalog number: 0 2195196 ); store at 4 °C
- Catalase (Fisher Scientific, catalog number: S25239A )
- Trolox (Acros Organics, catalog number: 218940050 )
- Sodium acetate (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9740 )
- Tricaine (Sigma-Aldrich, catalog number: E10521-10G ); store at -20 °C
- Vaseline
- Lanolin
- Paraffin
- Fixation: nuclease-free (see Recipes)
- 1x PBS + 0.1% Tween-20 (PBSTw)
- 1x PBSTw + 0.25 mM levamisole
- 1x PBSTw + 3.7% formaldehyde
- 1x PBS + 0.1% Triton X-100
- 70% ethanol
- Probe and hybridization: nuclease-free (see Recipes)
- TE buffer
- Formamide
- smFISH wash buffer
- smFISH wash buffer + DAPI
- Hybridization buffer (HB)
- TE buffer
- Reagents for GLOX buffer/mounting medium: nuclease-free (see Recipes)
- GLOX buffer, no enzymes
- GLOX buffer + enzymes
- 10% glucose
- Glucose oxidase, 3.7 mg/ml
- Catalase
- 200 mM Trolox
- VALAP
Equipment
- Pipettors (e.g., Mettler-Toledo, Rainin, model: P10, P20, P200, P1000)
- Glass Petri dish (e.g., diameter: 5 cm) or depression slide for dissecting worms
- Rotator (e.g., BD, Clay Adams Nutator, model: 421105 )
- Microcentrifuge at room temperature (Eppendorf, model: 5415 D )
- 37 °C incubator
- Vortexer (e.g., Baxter, catalog number: S8223-1 )
- Confocal microscope (Leica Microsystems, model: Leica TCS SP8 )
Procedure
Note: Figure 1 shows a flowchart of the protocol. All spins are at 400 x g (2,000 rpm) for 30 sec unless otherwise stated and buffer recipes are listed at the end.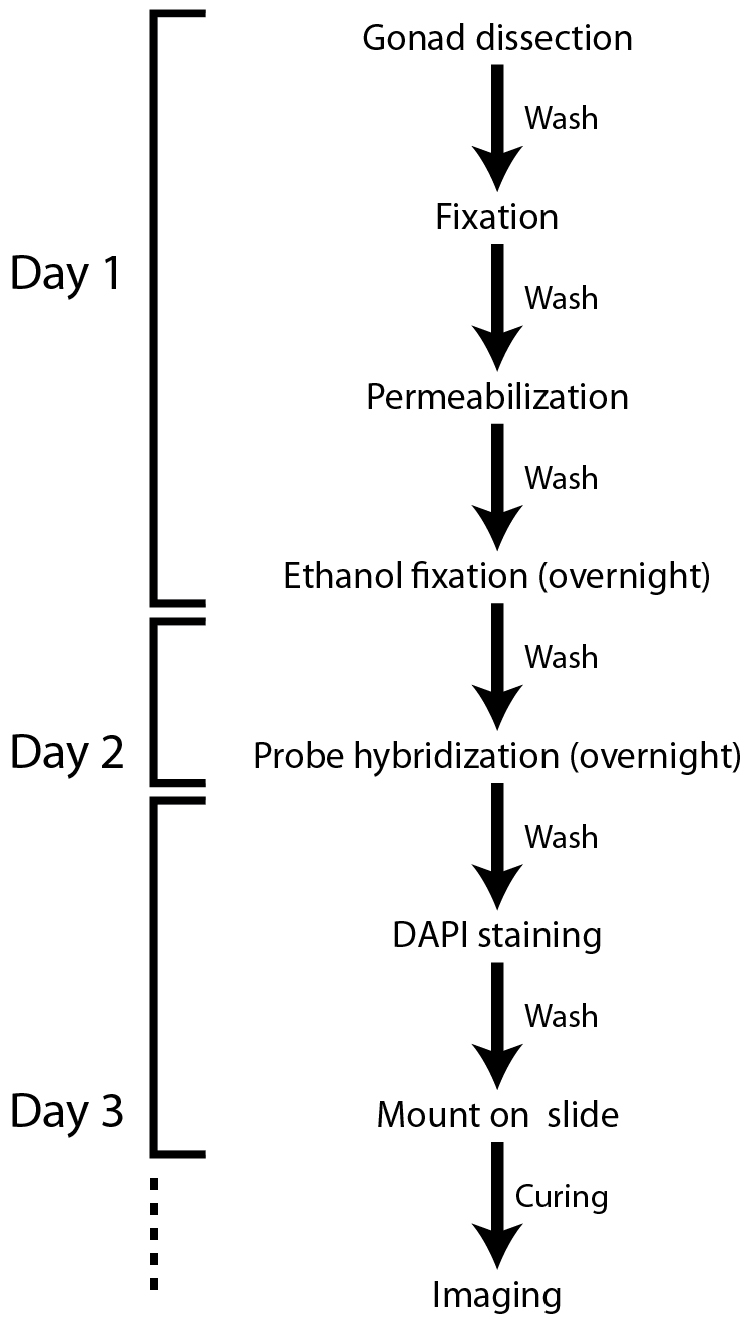
Figure 1. Flowchart of the smFISH protocol for the C. elegans gonad
- Probe preparation for use
smFISH probes can be designed and purchased through the Biosearch Technologies website (https://www.biosearchtech.com/). Choose fluorophores compatible with the microscopic system used for the experiments. For multiplexed probes, carefully choose probe-conjugated fluorophores to eliminate bleed-through between channels during image acquisition. To store smFISH probes, resuspend in 20 µl TE buffer (see Recipes) at pH 8.0 (final concentration of 250 µM) and store at -20 °C. This stock solution can be diluted to an appropriate dilution (e.g., 1:10-1:100) in TE buffer to make the working solution. Choose the dilution that works best for smFISH (e.g., the highest signal-to-noise ratio) and keep diluted probe solution at -20 °C in the dark. The frozen probe solution should be thawed at RT when used for smFISH. In our hands, multiple rounds of freezing-thawing did not affect the performance of smFISH. Do not boil the probes. - Day 1: Gonad dissections for smFISH
- Wipe down bench top, gloves, and pipettors with RNaseZap®.
- Pick worms from growth plates into glass Petri dish (or depression slide) with PBS with 0.1% Tween-20 (PBSTw) containing anesthesia (e.g., 0.25 mM levamisole [or tetramisole]). Alternatively, add 200-300 µl PBSTw with anesthesia to growth plate and pipet worms into Petri dish or depression slide. A larger volume (1-2 ml) of PBSTw can also be used for moving worms into Petri dish. In case of whole worm staining, skip steps B2-B3 and proceed to step B4.
- Extrude gonads as quickly as possible (< 10 min including spin) directly in the Petri dish. Cut behind pharynx or at tail with a #10 scalpel or a needle (Figure 2) (Crittenden et al., 2017).
- Pipet extruded gonads into a 1.5 ml microcentrifuge tube. Spin and remove supernatant.
- Add 1 ml fix solution (final 3.7% formaldehyde v/v in 1x PBS + 0.1% Tween-20) and incubate for at least 15 min (up to 45 min) at room temperature (RT) on rocker.
- Spin down the sample.
- Wash 1 x in 1 ml PBSTw.
- Incubate for 10 min in 1 ml 1x PBS + 0.1% Triton X-100 at RT to permeabilize.
- Wash 2 x with 1 ml PBSTw.
- Resuspend in 1 ml 70% EtOH and incubate at 4 °C for 16-18 h (overnight). The sample can be stored in 70% EtOH for up to one week.
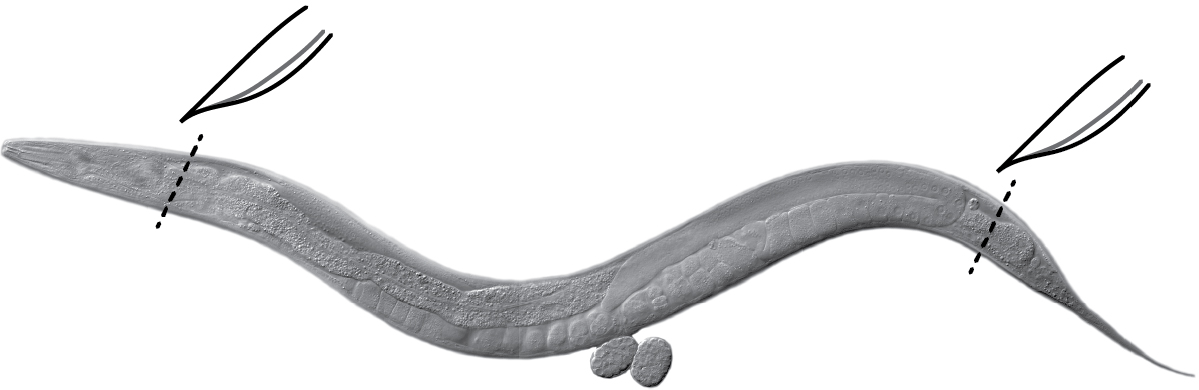
Figure 2. Gonad extrusion. Cut the head (behind the pharynx) or tail of the worm (black dashed lines) using a scalpel or a needle to extrude gonads. Internal pressure of the worm causes gonads to extrude readily. Image: Maria Gallegos.
- Wipe down bench top, gloves, and pipettors with RNaseZap®.
- Day 2: Hybridization
- Make wash buffer fresh, letting formamide come to RT before opening.
- Bring hybridization buffer (HB) to RT before opening.
- Thaw diluted probe, protecting from light.
- Spin the extruded gonads prepared on Day 1, remove ethanol.
- Add 1 ml wash buffer, equilibrate for 5 min at RT. Spin and remove as much buffer as possible.
- Add 1 μl diluted probe to 100 μl HB per sample (final probe concentration of 0.025-0.5 µM) and gently flick the tube (see Day 2 hybridization notes).
- Resuspend sample in HB with probe and mix well.
- Incubate overnight at 37 °C, with rotation and protected from light.
Note: You can leave samples for up to 48 h at this step.
- Make wash buffer fresh, letting formamide come to RT before opening.
- Day 3: Wash and mount samples (all at room temperature)
- Add 1 ml smFISH wash buffer, invert to mix. Spin and remove buffer.
Note: If worms don’t pellet well due to HB viscosity, this spin may be done at 400-500 x g (2,200-2,400 rpm) for 1.5 min. - Wash with 1 ml wash buffer + 1 μg/ml DAPI for 30 min on rotator (protected from light).
- Wash 2 x with 1 ml wash buffer.
Note: Either wash 5 min on the rotator or invert tube ~10 times before spinning down again. - Remove as much wash buffer as possible from fixed samples.
- Resuspend gonads in the mounting media (e.g., ProLong Gold Antifade Mountant or GLOX buffer).
- Let sit for 30 min to a few hours.
- Pipet 12-15 µl onto a slide, cover with a 22 x 22 coverslip. Remove excess liquid.
- Cure at least overnight and up to 4 days in the dark at room temp or 4 °C. The ProLong Gold user manual recommends a ‘curing’ process (~24 h at RT) for best performance. We have observed that the curing process does improve fluorescence signal.
- Seal with nail polish for long term storage.
- Add 1 ml smFISH wash buffer, invert to mix. Spin and remove buffer.
Data analysis
Representative smFISH image (Figure 3).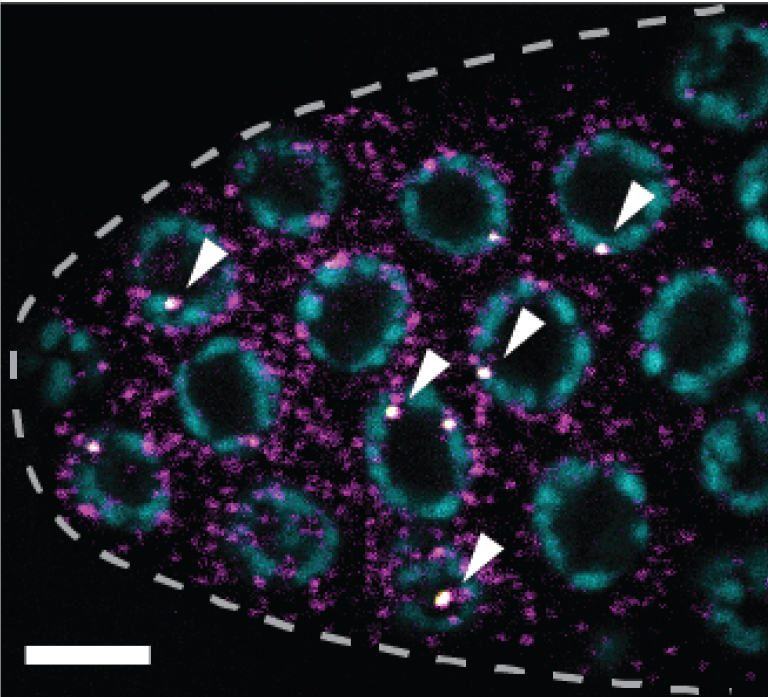
Figure 3. sygl-1 smFISH at the distal gonad. The sygl-1 gene is a direct target of Notch transcriptional activation. SYGL-1 protein is a critical regulator for germline stem cell (GSC) maintenance. Individual nascent transcripts (bright white nuclear spots; overlay of yellow and magenta channel, indicated by arrowheads) and cytoplasmic mRNA (magenta) of sygl-1 are visualized with spectrally distinct smFISH probes. DAPI marks DNA (blue). Maximum Z-projection is shown. Dashed line marks gonadal outline. Scale bar = 5 µm. Modified from Lee et al. (2016b).
Images taken using confocal microscopy do not require further image processing. However, images taken using wide-field (compound) microscopy must be deconvolved with carefully chosen image processing conditions (e.g., iterations). A caveat to the wide-field method is that over- or under-processed images can produce false-positive or false-negative signals. Using confocal images, the RNA spots can be systematically quantified with standard image processing (e.g., ImageJ) or published tools (e.g., MATLAB codes) (Mueller et al., 2013; Lee et al., 2016b). The wide-field microscope with a typical CCD camera (e.g., Hamamatsu C11440) generates microscopic images with the resolution high enough (pixel size: 0.1-0.2 µm) for identifying single RNA spots (Lee et al., 2016a). For confocal microscopy, we recommend using a pixel size similar to the size that works for wide-field microscopy (Abbaszadeh and Gavis, 2016; Lee et al., 2016b). As a first step of smFISH analysis, we quantify the size, shape (e.g., Gaussian distribution), signal intensities and subcellular location of detected RNA spots to validate singularity of detected RNA spots. In the Kimble lab, we consistently see nuclei containing one to four active transcription sites using intron-specific smFISH probes, with four matching the maximum number of loci of a gene (after DNA replication in S phase). The nascent RNAs are nuclear and colocalized to DAPI (or other DNA strain of choice). The mRNAs are cytoplasmic. Detected mRNA spots show tight signal distribution (C.V. < 0.5) in most RNA species, indicating that the detected spots are likely single RNA molecules (Lee et al., 2013 and 2016b). Control experiments such as detecting RNAs in the target gene knock-out background and using a different set of probes hybridizing the same target gene should be done to ensure the specificity of the probes to the target gene.
Notes
- Day 1, probe design notes
We used the standard settings of smFISH probe designer (Biosearch Technologies), which spread the probes throughout the gene. We have not tested whether smFISH performance changes depending on the region the probes bind, but one prediction is that 5’ probes will better visualize earlier transcription events. RNA-seq data from consortiums (e.g., ModEncode) may provide information on the gene expression level before choosing the target gene. For the C. elegans germline, we use RNA-seq data (Ortiz et al., 2014 and Wormbase), in-situ hybridization data (NEXTDB) and qPCR to estimate the level of target gene expression. - Day 1, dissection notes
Periodically re-wipe gloves and pipettors with RNaseZap®. Use RNase-free filtered tips and RNase-free tubes; keep tip boxes closed when not in active use. Put 50-100 worms in the same tube for the entire protocol. - Day 2, hybridization notes
- Minimize exposure to light during and after hybridization. Shield tube in your hand while changing buffers/tips, and wrap in foil for washes. To remove liquid from samples, use a P1000 pipettor because it can remove most liquid (within ~20 μl) without exposing tube to dissection scope light. The exceptions are before adding HB and before adding mounting media (look under the dissection scope [or check your tip before expelling liquid] and use a P1000, then a P200 and then a P10 to remove as much wash buffer as possible).
- Probe dilution is an effective way to optimize and should be determined empirically for each probe set. Probe stocks (250 μM) are diluted in RNase-free TE buffer before adding to the HB and typical final concentrations in HB are 0.025-0.5 μM (usually 0.25 μM is the first concentration we try). More dilute = less background.
- While samples are equilibrating in wash buffer, mix HB and probe in a separate RNase-free tube. It is recommended to use 50-100 μl HB + 1-3 µl probe per sample of fixed dissected worms.
Note: Adding HB and probe sequentially on top of fixed germlines can also work, but mix gently and thoroughly–no vortexing.
- Minimize exposure to light during and after hybridization. Shield tube in your hand while changing buffers/tips, and wrap in foil for washes. To remove liquid from samples, use a P1000 pipettor because it can remove most liquid (within ~20 μl) without exposing tube to dissection scope light. The exceptions are before adding HB and before adding mounting media (look under the dissection scope [or check your tip before expelling liquid] and use a P1000, then a P200 and then a P10 to remove as much wash buffer as possible).
- Day 3 notes
- Washing and mounting notes
Note: With the Quasar 570 probe, either ProLong Gold Antifade or GLOX mounting media can be used. We saw essentially no difference in signal quality between the two media. - Using ProLong Gold Antifade Mountant (PLG, Life Technologies):
- Slides and coverslips can be baked as an extra precaution against RNase contamination, or simply dedicate a box to smFISH.
- Collect any gonads/PLG remaining in tube and add to drop of gonads/PLG on the slide. Or if there is a lot left, mount on a separate slide.
- Remove bubbles from PLG droplet, being careful not to take gonads with them. Recommended methods include: popping with eyelash tool or flamed hot platinum wire, or aspirating gently with P10. Arrange worms inside drop quickly, using a pick to spread samples evenly.
- Cover with a 22 x 22 mm square coverslip. Place in drawer or other dark areas, on flat surface, at RT to cure. Put gentle pressure on coverslip to remove excess liquid and flatten gonads.
- Seal coverslip edges with nail polish. Stored in the dark at 4 °C, samples last for months.
- Using GLOX buffer for sensitive fluorophores (e.g., Quasar 570 and Quasar 670)
- Remove wash buffer, resuspend in 850 μl GLOX without enzymes. Let sit for 5 min (or place at 4 °C several hours until ready to image, as this method images each sample immediately after adding GLOX + enzymes).
- Immediately before imaging, remove buffer and add 100 μl GLOX + enzymes.
- Dissected gonads are extremely sticky; it is helpful to pipette up and down once in 1-10% Triton X-100 to coat the inside of the pipette.
- Pipette 4 μl sample onto a 22 x 22 mm square coverslip.
- Place a 12 x 12 mm square coverslip over sample drop. Lay a Kimwipe over the sample and press lightly on edges to remove excess liquid.
- Invert coverslip ‘sandwich’ so that the 12 x 12 coverslip is face down and placed on microscope slide to create imaging chamber. Seal with VALAP, starting with corners and then sealing edges. Be very careful that VALAP seal is airtight.
- Imaging notes
Note: In case of using multiple fluorophores, carefully select the dichroic mirrors (wide-field microscope) or acquisition wavelength range (confocal microscope) to avoid signal bleed-through. Use settings to minimize photobleaching for image analysis and single-molecule quantitation. We recommend acquiring images sequentially, channel by channel, starting from the excitation light with the longest wavelength for the best performance. Below are the general smFISH image acquisition settings used in the Kimble lab. - Microscope: Leica SP8 confocal
Objective: 63x 1.4
Zoom 300% to focus on distal ~60-70 µm of germline.
Frequency: 400 Hz
Laser power: between 1-5%, typically 3.5%
Gain: typically 40 for HyD detectors
8-16 line averaging, no accumulation
Pinhole: ~1 Airy unit. Match pinholes for exon and intron channels to improve colocalization. We have adjusted to between 1.0 and 1.5 to gather more light–typically, we stay below 1.25-1.3.
Bidirectional X: ON, phase = -32.40
Z-stack interval = 0.3 μm
Note: This way transcription sites are present in more than one slice and all mRNAs should be captured. - Identifying C. elegans gonads: The gonads are typically attached to the worm carcass but sometimes they fall off during the wash step. Both attached and detached gonads can be imaged for smFISH. The extruded gonad is an elongate light-colored tissue with hundreds of packed small cells. The extruded intestine, by contrast, is an elongate dark-colored tissue with only tens of large cells and is therefore morphologically different from the gonad. It is not necessary to isolate the gonads but the microscopic field can be adjusted so that only the region of interest within the gonad will be imaged.
- Washing and mounting notes
Recipes
- Fixation: nuclease-free
- 1x PBS + 0.1% Tween-20 (PBSTw) (need 5 ml per sample)
50 ml 1x PBS, nuclease-free
50 μl Tween-20 - 1x PBSTw + 0.25 mM levamisole
1 ml 1x PBS, nuclease-free
1 µl Tween-20
1 µl 0.25 M levamisole stock solution (51 mg in 1 ml M9 or 1x PBS) - 1x PBSTw + 3.7% formaldehyde (need 1 ml per sample, make fresh)
900 µl PBSTw
100 μl 37% formaldehyde - 1x PBS + 0.1% Triton X-100 (need 1 ml per sample)
9.9 ml 1x PBS, nuclease-free
100 μl 10% Triton X-100, diluted in nuclease-free water - 70% ethanol (need 1 ml per sample)
3 ml 100% EtOH, nuclease-free
7 ml H2O, nuclease-free
- 1x PBS + 0.1% Tween-20 (PBSTw) (need 5 ml per sample)
- Probe and hybridization: nuclease-free
- TE buffer (DEPC-treated)
10 mM Tris base
1 mM EDTA
pH 8.0 - Formamide
Aliquot and store at 4 °C, warm to RT before opening - smFISH wash buffer (need 6 ml per sample)
1 ml 20x SSC
1 ml formamide
8 ml DEPC water
10 μl Tween-20 - smFISH wash buffer + DAPI (need 1 ml per sample)
1 ml wash buffer
1 μl DAPI (1 mg/ml, stored in the dark at 4 °C) - Hybridization buffer (HB)
*Note: 10% formamide is generally used. However, formamide concentration can be adjusted to modify stringency. If target RNA has high GC content, try increasing formamide to 15-20%, or up to 50%. If adjusting formamide concentration, match concentration in wash buffer.
1 g dextran sulfate
7.3 ml DEPC water (or up to 10 ml volume)
1 ml 20x SSC
1 ml formamide
Notes:- Combine dextran sulfate and DEPC-H2O, rock ~30 min until fully dissolved. Add 20x SSC and formamide; invert to mix. Store in 500 μl aliquots at -20 °C.
- *Other labs (see for example Lee et al., 2013) also add (per 10 ml HB):
10 mg E. coli tRNA
100 μl Vanadyl ribonucleoside complex (200 mM)
40 μl BSA (nuclease-free) (50 mg/ml)
We started with these reagents but gradually omitted them. If experiencing problems with background, try adding them back in one at a time (in order listed).
- Combine dextran sulfate and DEPC-H2O, rock ~30 min until fully dissolved. Add 20x SSC and formamide; invert to mix. Store in 500 μl aliquots at -20 °C.
- TE buffer (DEPC-treated)
- Reagents for GLOX buffer/mounting medium: nuclease-free
- GLOX buffer, no enzymes (need 1 ml per sample)
850 µl H2O, nuclease-free
100 µl 20x SSC, nuclease-free
40 µl 10% glucose, nuclease-free
10 µl 1 M Tris, pH 8.0, nuclease-free [TE also works]
Note: Prepare fresh immediately before use. - GLOX buffer + enzymes
100 µl GLOX buffer
1 µl glucose oxidase, 3.7 mg/ml
1 µl catalase
1 µl 200 mM Trolox
Note: Keep on ice or at 4 °C. - 10% glucose
5 g glucose (weighed out in nuclease-free way)
50 ml nuclease-free H2O
Note: Pass through a 0.2 µm syringe filter and store at 4 °C. - Glucose oxidase, 3.7 mg/ml
37 mg glucose oxidase (weighed out in nuclease-free way)
167 µl 3 M sodium acetate, pH 5.5, nuclease-free
10 ml H2O, nuclease-free
Note: Divide into 100 µl aliquots and store at -20 °C. - Catalase
Note: Store in the dark at 4 °C. Vortex or pipet up and down before use but do not spin. - 200 mM Trolox
5 mg Trolox (weighed out in nuclease-free way)
1 ml 100% ethanol
Note: Store as 100 µl aliquots at -20 °C. - VALAP
Vaseline + lanolin + paraffin (1:1:1 w/w/w)
Note: To use, melt at ~70 °C and apply with paintbrush or metal spatula.
- GLOX buffer, no enzymes (need 1 ml per sample)
Acknowledgments
This protocol has been adapted from Lee et al. (2016b). ESK was supported by the American Cancer Society–George F. Hamel Jr. Fellowship (PF-14-147-01-DDC). HSS was supported by an Ellison Medical Foundation Fellowship of the Life Science Research Foundation. TRL is supported by the National Science Foundation Graduate Research Fellowship Program (Grant No. DGE-1256259). JK is an Investigator of the Howard Hughes Medical Institute.
References
- Abbaszadeh, E. K. and Gavis, E. R. (2016). Fixed and live visualization of RNAs in Drosophila oocytes and embryos. Methods 98: 34-41.
- Crittenden, S. L., Seidel, H. S. and Kimble, J. (2017). Analysis of the C. elegans germline stem cell pool. Methods Mol Biol 1463: 1-33.
- Ji, N. and van Oudenaarden, A. (2012). Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook: 1-16.
- Lee, C., Roberts, S. E. and Gladfelter, A. S. (2016a). Quantitative spatial analysis of transcripts in multinucleate cells using single-molecule FISH. Methods 98: 124-133.
- Lee, C., Sorensen, E. B., Lynch, T. R. and Kimble, J. (2016b). C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife 5.
- Lee, C., Zhang, H., Baker, A. E., Occhipinti, P., Borsuk, M. E. and Gladfelter, A. S. (2013). Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell 25(6): 572-584.
- Mueller, F., Senecal, A., Tantale, K., Marie-Nelly, H., Ly, N., Collin, O., Basyuk, E., Bertrand, E., Darzacq, X. and Zimmer, C. (2013). FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods 10(4): 277-278.
- Ortiz, M. A., Noble, D., Sorokin, E. P. and Kimble, J. (2014). A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765-1772.
- Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. and Tyagi, S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5(10): 877-879.
Article Information
Copyright
Lee et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Lee, C., Seidel, H. S., Lynch, T. R., Sorensen, E. B., Crittenden, S. L. and Kimble, J. (2017). Single-molecule RNA Fluorescence in situ Hybridization (smFISH) in Caenorhabditis elegans. Bio-protocol 7(12): e2357. DOI: 10.21769/BioProtoc.2357.
- Lee, C., Sorensen, E. B., Lynch, T. R. and Kimble, J. (2016b). C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife 5.
Category
Molecular Biology > RNA > RNA detection
Molecular Biology > RNA > Transcription
Cell Biology > Cell staining > Nucleic acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









