- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Semi-quantitative Analysis of H4K20me1 Levels in Living Cells Using Mintbody
Published: Vol 7, Iss 10, May 20, 2017 DOI: 10.21769/BioProtoc.2276 Views: 9968
Reviewed by: Ralph BottcherCody KimeMartin V Kolev

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Generation of Agarose-Based FFPE Cancer Organoids for Morphology Preservation
Mi Rim Lee [...] Yun-Hee Kim
Oct 5, 2025 1703 Views

Improved Immunohistochemistry of Mouse Eye Sections Using Davidson's Fixative and Melanin Bleaching
Anne Nathalie Longakit [...] Catherine D. Van Raamsdonk
Nov 20, 2025 1565 Views

Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Anastasia Yerushkin [...] Amir Dori
Dec 5, 2025 1224 Views
Abstract
Eukaryotic nuclear DNA wraps around histone proteins to form a nucleosome, a basic unit of chromatin. Posttranslational modification of histones plays an important role in gene regulation and chromosome duplication. Some modifications are quite stable to be an epigenetic memory, and others exhibit rapid turnover or fluctuate during the cell cycle. Histone H4 Lys20 monomethylation (H4K20me1) has been shown to be involved in chromosome condensation, segregation, replication and repair. H4K20 methylation is controlled through a few methyltransferases, PR-Set7/Set8, SUV420H1, and SUV420H2, and a demethylase, PHF8. In cycling cells, the level of H4K20me1 increases during G2 and M phases and decreases during G1 phase. To monitor the local concentration and global fluctuation of histone modifications in living cells, we have developed a genetically encoded probe termed mintbody (modification-specific intracellular antibody; Sato et al., 2013 and 2016). By measuring the nuclear to cytoplasmic intensity ratio, the relative level of H4K20me1 in individual cells can be monitored. This detailed protocol allows the semi-quantitative analysis of the effects of methyltransferases on H4K20me1 levels in living cells based on H4K20me1-mintbody described by Sato et al. (2016).
Keywords: Post translational modificationBackground
Posttranslational modifications of histone proteins play important roles in transcriptional regulation and genome integrity. While the one-dimensional epigenomic landscape has been revealed in many cell types by chromatin immunoprecipitation and sequencing, less is known about the dynamics of histone modifications due to technical limitations (Kimura et al., 2015). Recently, a few techniques for detecting protein modifications in living cells have been developed. One strategy uses sensors based on fluorescence/Förster resonance energy transfer (FRET) to monitor the balance between the modifying and demodifying enzymes. However, the dynamics of endogenous modifications cannot be monitored using FRET sensors. Another strategy that we have developed uses probes based on modification-specific antibodies. Fab-based live endogenous modification labeling (FabLEM) is a live-imaging system using fluorescently labeled antigen-binding fragments (Fabs). Fabs loaded into cells bind to the target modification without disturbing cell function as the binding time is very small (a second to tens of seconds). A genetically encoded system to express a modification-specific intracellular antibody (mintbody) can be applied for observation with a longer period of time or in living animals (Figure 1). Both Fabs and mintbodies are just small enough to pass through the nuclear pore by diffusion. When the level of the target modification increases, more probes become enriched in the nucleus. Therefore, by measuring the nuclear/cytoplasmic intensity ratio, changes of modification level in living cells can be monitored (Hayashi-Takanaka et al., 2011; Sato et al., 2013 and 2016). The live cell modification monitoring system using mintbodies will be particularly useful to evaluate the effects of small chemicals and protein depletion and overexpression.
Histone H4 Lys20 monomethylation (H4K20me1) is an essential modification in mammals, involved in chromosome condensation, segregation, replication and repair, as well as gene regulation (Beck et al., 2012; Jørgensen et al., 2013). The level of H4K20me1 increases during G2 to M phases and the inhibition of PR-Set7/Set8, a methyltransferase responsible for H4K20 monomethylation, causes mitotic defects. In female cells, the enrichment of H4K20me1 in inactive X chromosomes is microscopically observed. H4K20me1-specific mintbody has proven useful for monitoring the dynamic behavior of H4K20me1 in living cells (Sato et al., 2016). In addition, alteration of H4K20me1 level by ectopic expression of a methyltransferase has been evaluated. Among methyltransferases (PR-Set7/Set8, SUV420H1, and SUV420H2) and a demethylase (PHF8), involved in H4K20me1 metabolism, the expression of SUV420H1, which add methyl-groups to monomethylated H4K20 towards to trimethylation, caused a drastic effect. As an example of measuring relative H4K20me1 levels, we here describe the method to evaluate the effect of SUV420H1 on H4K20me1 in living cells.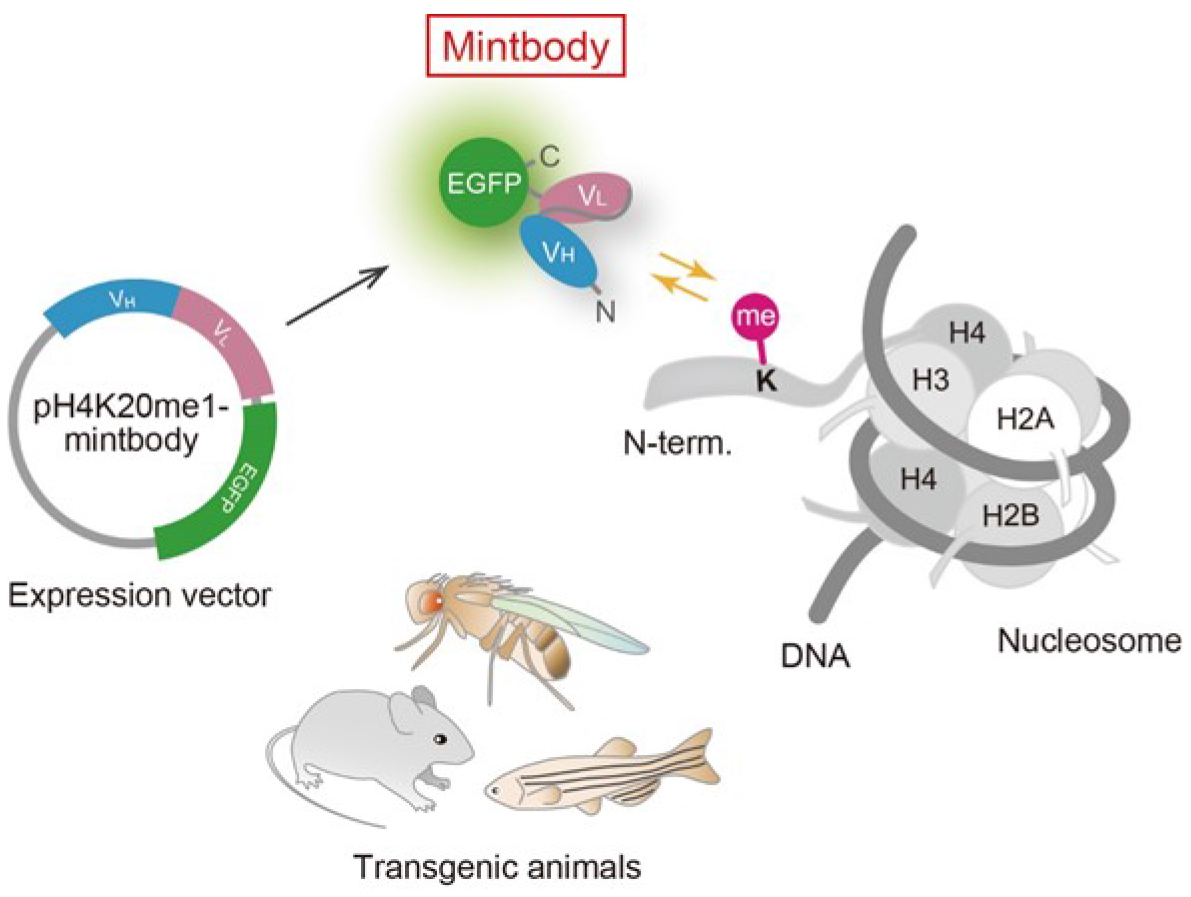
Figure 1. Schematic diagram of mintbody expression and function. A genetically encoded mintbody, which reversibly binds to specific modification, can be expressed in cells and animals that harbors the expression vector.
Materials and Reagents
- Pipette tips (10, 20, 200, 1,000 μl)
- 6 well plate (Corning, catalog number: 3516 )
- 10 cm dish (Greiner Bio One International, catalog number: 664160-013 )
- 24 well glass-bottom plate (IWAKI, catalog number: 5826-024 )
- HeLa cells (ATCC, catalog number: CRM-CCL-2 )
- Purified plasmid DNA (~1 μg/μl) encoding H4K20me1-mintbody based on pEGFP (Clontech) or a piggybac system (Sato et al., 2016) and Halo-SUV420H1 (Kazusa DNA Research Institute; FlexiHaloTag clone FHC01413)
- Dulbecco’s modified Eagle’s medium (DMEM), high glucose (4 g/L), containing L-Gln and sodium pyruvate (Nacalai Tesque, catalog number: 08458-16 )
- L-glutamine-penicillin-streptomycin solution (Sigma-Aldrich, catalog number: G1146-100ML )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- FuGENE HD (Promega, catalog number: E2312 )
- Opti-MEM media (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- G-418 disulfate aqueous solution (Nacalai Tesque, catalog number: 16513-26 )
- FluoroBrite DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: A1896701 )
- HaloTag TMR ligand (Promega, catalog number: G8251 )
Notes:
- Nucleotide sequence of H4K20me1-scFv is available in public databases (DDBJ/EMBL/GenBank) under the accession number LC129890. The plasmid DNA is available upon request to the authors.
- The original H4K20me1-specific antibody that was used to generate H4K20me1-mintbody (Hayashi-Takanaka et al., 2015) is available commercially (MBL International, catalog number: MABI0421).
Equipment
- CO2 incubator
- Pipette (10, 20, 200, 1,000 μl)
- 35 mm glass-bottom dishes, No. 1.5 coverslip (MATTEK, catalog number: P35G-1.5-14-C )
- Fluorescence microscope (Nikon Instruments, model: ECLIPSE Ti-E ) operated by NIS-elements and equipped with:
A spinning disk confocal unit (Yokogawa Electric, model: CSU-W1 )
An EM-CCD camera (Andor, model: iXon3 DU888 X-8465 )
An objective lens (Plan Apo 40x DIC M N2 [NA 0.95])
A laser unit (Nikon Instruments, model: LU-N4 )
A heated stage (Tokai Hit)
A CO2-control system (Tokken)
Software
- NIS-elements ver. 4.30 (Nikon Instruments)
- Excel (Microsoft)
Procedure
- Establishing stable cell lines expressing the H4K20me1-mintbody (Figure 2)
Note: When using primary cells, cell sorting can be used instead of establishing antibiotic resistant clones.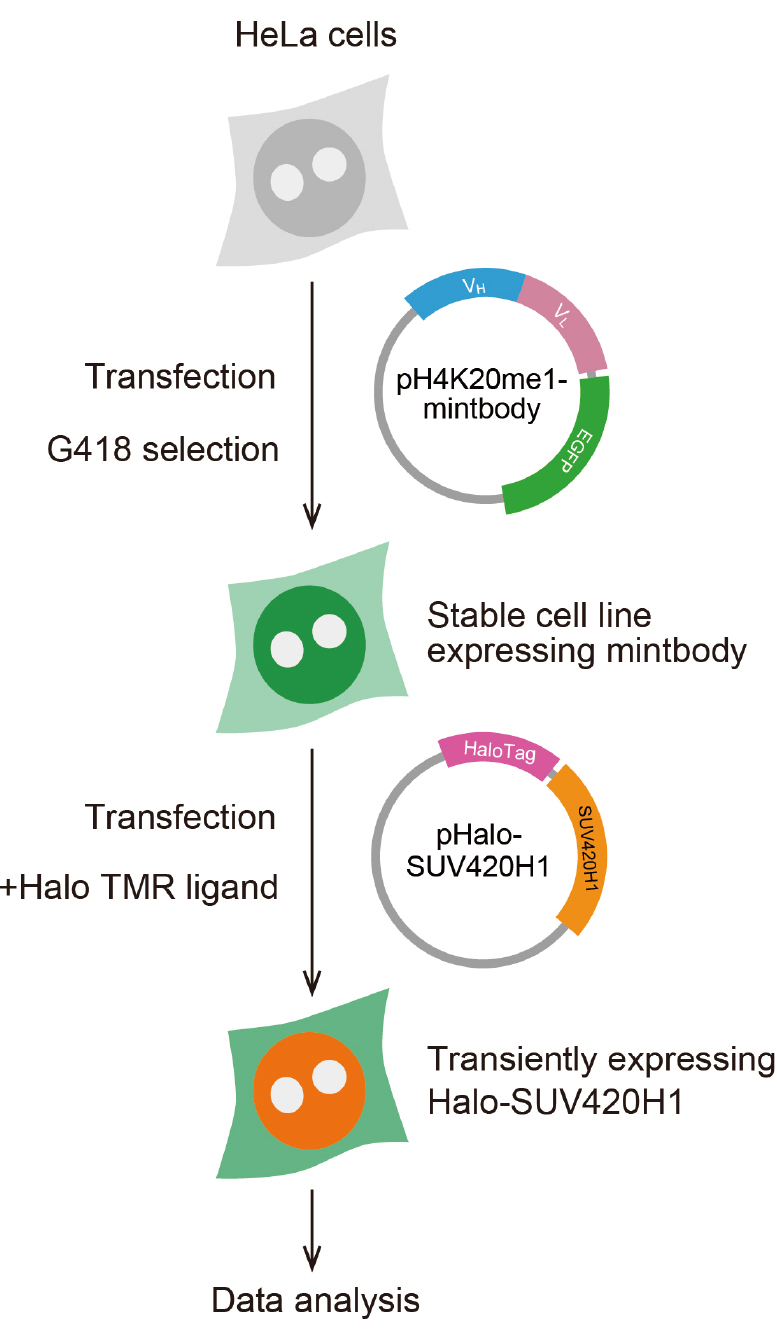
Figure 2. Scheme for preparing cells expressing H4K20me1-mintbody and Halo-SUV420H1- Maintain HeLa cells in DMEM supplemented with 10% FBS and L-glutamine-penicillin-streptomycin at 37 °C under 5% CO2 atmosphere, according to a standard protocol (Freshney, 2015).
- Transfect the plasmid DNA encoding the H4K20me1-mintbody into cells using FuGENE HD, as per standard protocol. Typically, cells were plated onto 6 well plate (2 x 105 cells in 2 ml medium/well) on the day before transfection. Mix 2 μg plasmid and 6 μl FuGENE HD in 100 μl Opti-MEM, leave the mixture for 5 min at room temperature, and add the mixture to one well of a 6-well plate containing 2 ml medium. Put back the plate into a CO2 incubator.
- The next day, to obtain the stable cell line expressing the mintbody, harvest the cells, dilute at around 103 cells/ml, and plate cells on to a 6 well plate (2 ml per well) or a 10 cm dish (10 ml per dish) in DMEM with FBS and L-glutamine-penicillin-streptomycin, containing 1 mg/ml G-418.
- Approximately two weeks after addition of G-418, isolate and expand GFP-positive colonies. Isolation of cells expressing H4K20me1-mintbody can be done in several ways (Freshney, 2015).
- If fluorescent colonies are observed at a high frequency (> 10%), randomly pick up 24 colonies using a micropipette with a 200 μl tip, or using a cloning ring, and transfer into a 24-well glass-bottom plate containing 0.5 ml DMEM containing FBS and L-glutamine-penicillin-streptomycin (without G-418).
- If fluorescent colonies are rarely observed, pick up them after marking their position under an inverted fluorescence microscope. A few days later, investigate the level and distribution of fluorescence in each well under an inverted fluorescence microscope using a dry lens with a high numerical aperture and a short working distance (e.g., Plan Apo 40x DIC M N2 [NA 0.95]). Fluorescent cells can also be collected using a cell sorter, if available.
- Maintain HeLa cells in DMEM supplemented with 10% FBS and L-glutamine-penicillin-streptomycin at 37 °C under 5% CO2 atmosphere, according to a standard protocol (Freshney, 2015).
- Cell preparation for imaging
- Two days before imaging, plate cells expressing mintbody in a 35 mm glass-bottom dish (2 ml medium; ~2 x 105 cells/dish).
- The next day, transfect the plasmid DNA encoding Halo-SUV420H1 into cells using FuGENE HD to convert monomethylated H4K20 to trimethylation. Transfection using 2 μg plasmid and 6 μl FuGENE HD is carried out as indicated above. The transaction efficiency typically yields ~30%.
- A day after transfection, replace the culture medium to FluoroBrite DMEM with 10% FBS and L-glutamine-penicillin-streptomycin, containing 0.1 μM Halo TMR ligand. Incubate the cells at 37 °C under 5% CO2 atmosphere.
- Two days before imaging, plate cells expressing mintbody in a 35 mm glass-bottom dish (2 ml medium; ~2 x 105 cells/dish).
- Image acquisition
- One hour after the addition of Halo TMR ligand, set the glass-bottom dish onto a heated stage. It is not essential to remove free TMR. However, if the expression level of HaloTag-tagged protein is low and background fluorescence hampers the detection, cells should be washed three times with FluoroBrite DMEM with 10% FBS and L-glutamine-penicillin-streptomycin, before setting the dish onto a heated stage.
- Acquire fluorescence images for the H4K20me1-mintbody (green channel) and TMR-labeled Halo-SUV420H1 (red channel) using a confocal microscope with laser lines at 488 and 561 nm, combined with a dichroic mirror DM405/488/561/640 and emission filters 520/30 and 617/73, respectively.
- One hour after the addition of Halo TMR ligand, set the glass-bottom dish onto a heated stage. It is not essential to remove free TMR. However, if the expression level of HaloTag-tagged protein is low and background fluorescence hampers the detection, cells should be washed three times with FluoroBrite DMEM with 10% FBS and L-glutamine-penicillin-streptomycin, before setting the dish onto a heated stage.
Notes:
- One hour staining with 0.1 μM Halo TMR ligand is usually sufficient to label HaloTag-tagged proteins. However, if the signal intensity is not high enough, samples can be incubated overnight with the TMR ligand. Washing with FluoroBrite DMEM with 10% FBS and L-glutamine-penicillin-streptomycin also improves the signal-to-noise ratio.
- To avoid the saturation of fluorescence intensity, we briefly scan multiple areas and set up the laser power and exposure time to cover all cells (except dead ones, which sometimes exhibit brightest fluorescence) are within the dynamic range of the camera for both green and red channels.
- Non-transfected cells in the captured images will be used for negative control cells.
- Empirically, at least 30 Halo-positive cells should be imaged for a good statistical analysis. Total 200 cells including non-transfected cells are typically imaged.
Data analysis
- The relative level of H4K20me1 measured by the nuclear enrichment of mintbody
Note: To compare the nuclear enrichment of H4K20me1-mintbody in different cells, measure the nucleus/cytoplasm intensity ratio in each cell.- Open the images using NIS-elements.
- Select the background ROI by choosing an area without cells.
- Obtain the net intensity images by background subtraction.
- Draw ROIs for the cell and nucleus manually using ‘Draw Polygonal ROI’ tool (Figure 3). Nuclear region can be selected automatically by thresholding and binarization, but the selecting cell region is often difficult due to the low signal intensity in the cytoplasm. For analysis of tens of cells, manual drawing is often quicker and accurate; as the selected areas are quite large, a difference in a few pixels do not affect much.
- Measure the mean intensities [I], areas [A], and total intensity [T = I x A] of the ROIs in green channel.
- Export the data to Excel.
- Open the exported data in Excel and calculate the intensity of the cytoplasm by the following equation: [I]cytoplasm = ([T]cell - [T]nucleus)/([A]cell - [A]nucleus).
- Calculate the nucleus/cytoplasm intensity ratio by dividing the mean intensity of the nucleus with that of cytoplasm (i.e., the ratio = [I]nucleus/[I]cytoplasm).
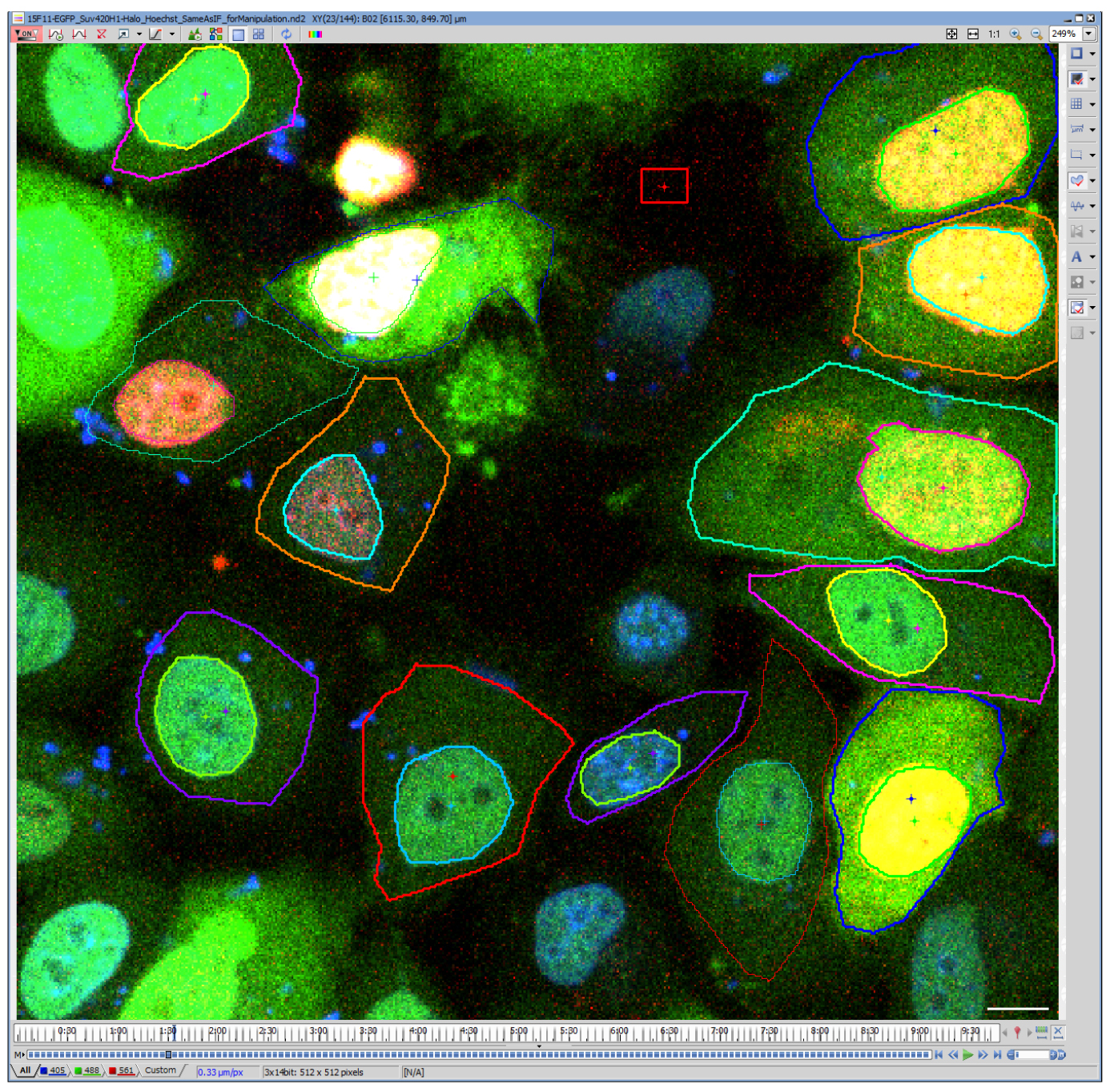
Figure 3. Drawing ROIs to define cell and nuclear regions. A representative image file analyzed using NIS-elements. Blue, Hoechst; Green, H4K20me1-mintbody; Red, Halo-SUV420H1. Note that live Hoechst staining was employed here, but this is not essential because the nuclear regions are clearly distinguished from the cytoplasm. Scale bar = 10 μm.
- Open the images using NIS-elements.
- The expression level of Halo-SUV420H1
- The ROI area of the nucleus defined above can also be used for the relative quantification of Halo-SUV420H1 expression level.
- Measure the mean intensity of the nucleus in red channel.
- The ROI area of the nucleus defined above can also be used for the relative quantification of Halo-SUV420H1 expression level.
- Relationship between SUV420H1 expression and H4K20me1
- Draw a scatter plot to visualize the relationship between the levels of SUV420H1 expression and H4K20me1 (Figure 4).
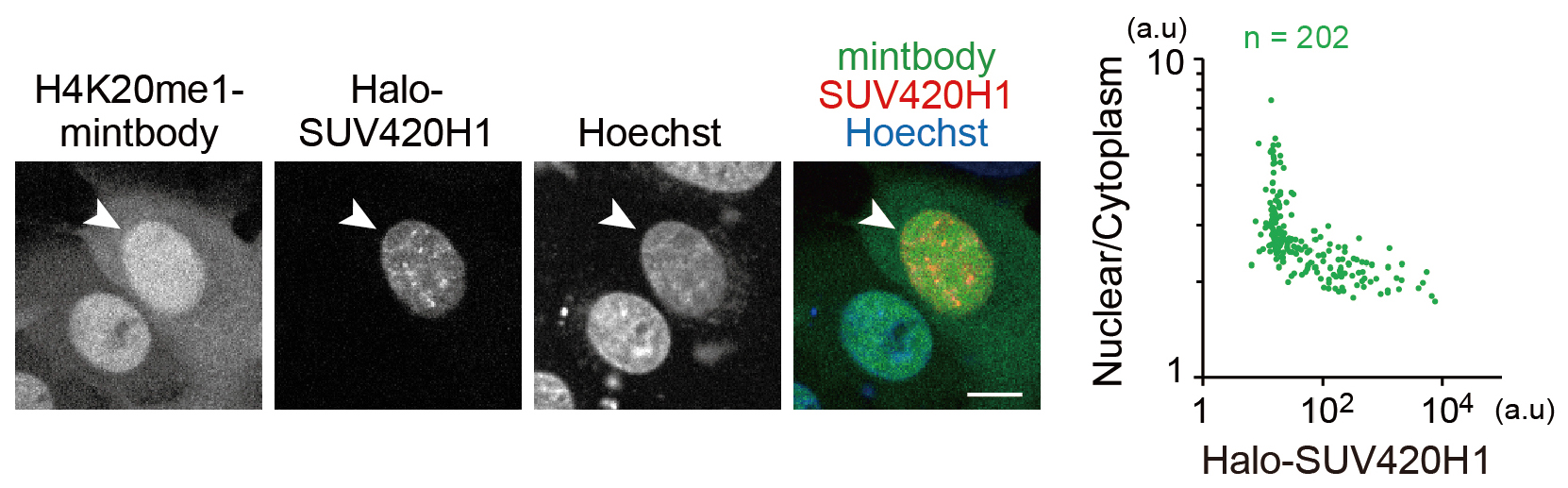
Figure 4. H4K20me1 levels in living cells monitored with H4K20me1-mintbody. This figure is adapted from Sato et al., 2016. Halo-SUV420H1 was transiently expressed in HeLa cells that stably express H4K20me1-mintbody. The mintbody was more diffuse in the cytoplasm in the cell that expressed SUV420H1 (arrowhead). Nuclear/Cytoplasm intensity ratios in single cells were plotted (right graph). Nuclear/Cytoplasm ratios decreased depending on Halo-SUV420H1 expression. Bar = 10 µm.
- Draw a scatter plot to visualize the relationship between the levels of SUV420H1 expression and H4K20me1 (Figure 4).
Notes
This protocol can be applicable to many other cell lines and modification enzymes. To confirm the live cell observations, immunofluorescence analysis based on fixed cells can be employed using the original H4K20me1-specific antibody that is used to generate H4K20me1-mintbody (Hayashi-Takanaka et al., 2015; Sato et al., 2016).
Acknowledgments
This work was supported by JSPS KAKENHI Grants JP25118714, JP262910711, JP25116005. The protocol has been adapted from Sato et al. (2016).
References
- Beck, D. B., Oda, H., Shen, S. S. and Reinberg, D. (2012). PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26(4): 325-337.
- Freshney, R. I. (2015). Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, 7th Edition. John Wiley & Sons.
- Hayashi-Takanaka, Y., Maehara, K., Harada, A., Umehara, T., Yokoyama, S., Obuse, C., Ohkawa, Y., Nozaki, N., Kimura, H. (2015). Distribution of histone H4 modifications as revealed by a panel of specific monoclonal antibodies. Chromosome Res 23(4):753-766.
- Hayashi-Takanaka, Y., Yamagata, K., Wakayama, T., Stasevich, T. J., Kainuma, T., Tsurimoto, T., Tachibana, M., Shinkai, Y., Kurumizaka, H., Nozaki, N., Kimura, H. (2011). Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res 39(15): 6475-6488.
- Jørgensen, S., Schotta, G. and Sorensen, C. S. (2013). Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 41(5): 2797-2806.
- Kimura, H., Hayashi-Takanaka, Y., Stasevich, T. J., Sato, Y. (2015). Visualizing posttranslational and epigenetic modifications of endogenous proteins in vivo. Histochem Cell Biol 144(2): 101-109.
- Sato, Y., Kujirai, T., Arai, R., Asakawa, H., Ohtsuki, C., Horikoshi, N., Yamagata, K., Ueda, J., Nagase, T., Haraguchi, T., Hiraoka, Y., Kimura, A., Kurumizaka, H. and Kimura, H. (2016). A genetically encoded probe for live-cell imaging of H4K20 monomethylation. J Mol Biol 428(20): 3885-3902.
- Sato, Y., Mukai, M., Ueda, J., Muraki, M., Stasevich, T. J., Horikoshi, N., Kujirai, T., Kita, H., Kimura, T., Hira, S., Okada, Y., Hayashi-Takanaka, Y., Obuse, C., Kurumizaka, H., Kawahara, A., Yamagata, K., Nozaki, N. and Kimura, H. (2013). Genetically encoded system to track histone modification in vivo. Sci Rep 3: 2436.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sato, Y. and Kimura, H. (2017). Semi-quantitative Analysis of H4K20me1 Levels in Living Cells Using Mintbody. Bio-protocol 7(10): e2276. DOI: 10.21769/BioProtoc.2276.
Category
Cancer Biology > General technique > Cell biology assays
Cell Biology > Cell staining > Protein
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









