- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay to Measure Interactions between Purified Drp1 and Synthetic Liposomes
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2266 Views: 11097
Reviewed by: Nicoletta CordaniJianwei SunDaniel Kraus

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phosphoinositides Coated Beads Binding Assay
Manuel Gálvez-Santisteban [...] Fernando Martin-Belmonte
Feb 5, 2014 12772 Views

Protein-lipid Interaction Analysis by Surface Plasmon Resonance (SPR)
Olga Lucia Baron and David Pauron
Sep 20, 2014 14554 Views

Competitive ELISA for Protein-lipopolysaccharide (LPS) Binding
Victoria Martínez-Sernández and Florencio M. Ubeira
Nov 20, 2014 15657 Views
Abstract
A mitochondrion is a dynamic intracellular organelle that actively divides and fuses to control its size, number and shape in cells. A regulated balance between mitochondrial division and fusion is fundamental to the function, distribution and turnover of mitochondria (Roy et al., 2015). Mitochondrial division is mediated by dynamin-related protein 1 (Drp1), a mechano-chemical GTPase that constricts mitochondrial membranes (Tamura et al., 2011). Mitochondrial membrane lipids such as phosphatidic acid and cardiolipin bind Drp1, and Drp1-phospholipid interactions provide key regulatory mechanisms for mitochondrial division (Montessuit et al., 2010; Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014; Stepanyants et al., 2015; Adachi et al., 2016). Here, we describe biochemical experiments that quantitatively measure interactions of Drp1 with lipids using purified recombinant Drp1 and synthetic liposomes with a defined set of phospholipids. This assay makes it possible to define the specificity of protein-lipid interaction and the role of the head group and acyl chains.
Keywords: MitochondriaBackground
Interactions of proteins and membrane lipids are critical for the remodeling of biological membranes in cells such as organelle division. In mitochondrial division, Drp1 constricts the mitochondrial membranes and drives this membrane remodeling process. We have recently shown that a signaling phospholipid, phosphatidic acid, interacts with Drp1 and creates the priming step by restraining assembled division machinery on mitochondria (Adachi et al., 2016). Drp1 recognizes both the head group and acyl chains of phosphatidic acid. To analyze Drp1-phosphatidic acid binding, we set up several protein-lipid interaction assays, including a lipid dot blot assay, a competition assay and a liposome flotation assay. These assays allowed us to determine the function of the head group and acyl chain composition in protein-lipid interactions. In addition, we analyzed lipids under bilayer conditions in the liposome assay and lipids under non-bilayer conditions in dot blot and competition assays, which made it possible to examine the contribution of the membrane curvature and lipid packing to protein-lipid interactions. Below, we describe a liposome flotation assay that can be applied to many peripheral membrane proteins.
Materials and Reagents
- 0.45 µm Millex-HA syringe filter unit (EMD Millipore, catalog number: SLHA033SS )
- 15-ml column (Poly-prep chromatography column) (Bio-Rad Laboratories, catalog number: 7311550 )
- Amicon Ultra-15 centrifugal filter unit with Ultracel-10 membrane (10k filter for domains of Drp1) (EMD Millipore, catalog number: UFC901024 )
- Amicon Ultra-15 centrifugal filter unit with Ultracel-50 membrane (50k filter for full length Drp1) (EMD Millipore, catalog number: UFC905024 )
- Polycarbonate membranes 0.4 μm (Avanti Polar Lipids, catalog number: 610007 )
- 96 well assay plate (Corning, catalog number: 3915 )
- Extruder set with holder/heating block (Avanti Polar Lipids, catalog number: 610000 )
- Filter support (Avanti Polar Lipids, catalog number: 610014 )
- 2 ml amber glass sample vials (WHEATON, catalog number: 224981 )
- Disposable culture tubes (Fisher Scientific, catalog number: 14-961-27 )
- 50 Ulrta-Clear tubes (Beckman Coulter, catalog number: 344057 )
- RosettaTM 2(DE3)pLysS competent cells (EMD Millipore, catalog number: 71403 )
- pET15b vectors (EMD Millipore, Novagen)
- 50% Ni-NTA beads (EMD Millipore, catalog number: 70666 )
- 4-20% Tris-HCl precast gel (Bio-Rad Laboratories, catalog number: 3450034 )
- Chloroform (Sigma-Aldrich, catalog number: C2432 )
- Ethanol (PHARMCO-AAPER, catalog number: 111000200 )
- DMSO (Fisher Scientific, catalog number: S369-500 )
- Liquid nitrogen
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids, catalog number: 850457 )
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (rhodamine-DPPE) (Avanti Polar Lipids, catalog number: 810158 )
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) (Avanti Polar Lipids, catalog number: 850705 )
- 1,2-dipalmitoyl-sn-glycero-3-phosphate (DPPA) (Avanti Polar Lipids, catalog number: 830855 )
- 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA) (Avanti Polar Lipids, catalog number: 840875 )
- 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphate (Palmitoyl lysoPA) (Avanti Polar Lipids, catalog number: 857123 )
- 1,2-dihexanoyl-sn-glycero-3-phosphate (Dihexanoyl PA) (Avanti Polar Lipids, catalog number: 830841 )
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) (Avanti Polar Lipids, catalog number: 850355 )
- 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids, catalog number: 850375 )
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) (Avanti Polar Lipids, catalog number: 850725 )
- 1,2-dipalmitoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (DPPG) (Avanti Polar Lipids, catalog number: 840455 )
- 1,2-dioleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (DOPG) (Avanti Polar Lipids, catalog number: 840475 )
- 1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (DPPS) (Avanti Polar Lipids, catalog number: 840037 )
- 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) (Avanti Polar Lipids, catalog number: 840035 )
- 1’,3’-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (TOCL) (Avanti Polar Lipids, catalog number: 710335 )
- 1’,3’-bis[1,2-dipalmitoyl-sn-glycero-3-phospho]-sn-glycerol TPCL (Echelon Biosciences, catalog number: L-C160 )
- Gas nitrogen
- SilverQuestTM Silver Staining Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: LC6070 )
- BactoTM tryptone (BD, BactoTM, catalog number: 211705 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271 )
- BactoTM yeast extract (BD, BactoTM, catalog number: 212750 )
- Agar (Alfa Aesar, catalog number: J10906 )
- Ultrapure water (e.g., MilliQ)
- Ampicillin, sodium salt (Alfa Aesar, catalog number: J11259 )
- Chloramphenicol (Mediatech, catalog number: 61-239-RI )
- beta-D-Galactopyranoside, Isopropyl-beta-D-thiogalactopyranoside (IPTG) (AMRESCO, catalog number: 0487 )
- HEPES (Fisher Scientific, catalog number: BP310-1 )
- Potassium hydroxide (KOH) (Avantor® Performance Materials, J.T. Baker, catalog number: 3116-01 )
- Magnesium chloride hydrate (MgCl2·6H2O) (Avantor® Performance Materials, J.T. Baker, catalog number: 2444 )
- Imidazole (Sigma-Aldrich, catalog number: I0125 or I202 )
Note: The product “ I0125 ” has been discontinue - 2-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Sodium phosphate monobasic (NaH2PO4·H2O) (Fisher Scientific, catalog number: S369-500 )
- Sodium phosphate dibasic heptahydrate (Na2HPO4·7H2O) (Fisher Scientific, catalog number: S373-3 )
- Sodium hydroxide (NaOH) (Avantor® Performance Materials, J.T. Baker, catalog number: 3722 )
- Coomassie Brilliant Blue R-250 (AMRESCO, catalog number: M128-50G )
- Acetic acid (Fisher Scientific, catalog number: A38-212 )
- Methanol (Fisher Scientific, catalog number: A412 )
- MES (2-morpholinoethanesulfonic acid, mono hydrate) (Sigma-Aldrich, catalog number: M8250 )
- Sucrose (Fisher Scientific, catalog number: S5-3 )
- LB plate containing ampicillin and chloramphenicol (see Recipes)
- LB with ampicillin and chloramphenicol (see Recipes)
- 0.5 M IPTG
- 0.5 M HEPES (pH 7.4) (see Recipes)
- Lysis buffer (see Recipes)
- Wash buffer (see Recipes)
- Elution buffer (see Recipes)
- 10x PBS (see Recipes)
- Coomassie Brilliant Blue solution (see Recipes)
- 100 mM MES (pH 7.0) (see Recipes)
- 20 mM MES (pH 7.0) (see Recipes)
- 20 mM MES (pH 7.0)/100 mM NaCl (see Recipes)
- 2.7 M sucrose/20 mM MES/100 mM NaCl (see Recipes)
- 1.25 M sucrose/20 mM MES/100 mM NaCl (see Recipes)
- 0.25 M sucrose/20 mM MES/100 mM NaCl (see Recipes)
Equipment
- 1 ml syringe (Hamilton, catalog number: 81365 )
- 500 µl syringe (Hamilton, catalog number: 81265 )
- 100 µl syringe (Hamilton, catalog number: 81065 )
- 50 µl syringe (Hamilton, catalog number: 80500 )
- 25 µl syringe (Hamilton, catalog number: 80400 )
- Incubator shaker (37 °C) (Eppendorf, New BrunswickTM, model: Series 25 )
- Incubator shaker (16 °C) (Lab Companion, model: IS-971R )
- 1,000 ml bottle (Beckman Coulter, catalog number: A98814 )
- JLA8.1000 rotor (Beckman Coulter, catalog number: 969329 )
- GH3.8 rotor (Beckman Coulter, catalog number: 360581 )
- Fisher Scientific Sonic dismembrator (Fisher Scientific, model: 100 )
- CS-6R centrifuge (Beckman Coulter, model: CS-6R )
- Avanti J-E (Beckman Coulter, model: Avanti J-E , catalog number: 369001)
- SpeedVac SVC100 (Savant System, model: SpeedVac SVC100 )
- Heat block (37 °C, 42 °C, 95 °C) (Fisher Scientific, model: Fisher ScientificTM IsotempTM Digital and Analog Dry Bath Incubators , catalog number: 11-718)
- Motorized upright fluorescence microscope (Olympus, model: BX61 )
- POLARstar Omega (BMG LABTECH, model: POLARstar Omega )
- Labquake Shaker Rotisserie (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4152110 )
- SW55Ti rotor (Beckman Coulter, catalog number: 342196 )
- Avanti J-26XP (Beckman Coulter, model: Avanti® J-26 XP , catalog number: 393124)
- Optima L-100XP ultracentrifuge (Beckman Coulter, model: OptimaTM L-100XP , catalog number: 392050)
- Scanmaker 8700 (Microtek, model: Scanmaker 8700 )
- Power supply (Bio-Rad Laboratories, model: PowerPacTM HC )
- Multi-Purpose rotator (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 2309 )
- Vortex mixer (Fisher Scientific, model: Fisher ScientificTM Analog Vortex Mixer , catalog number: 02215365)
Software
- ImageJ (Version 1.48, http://imagej.net/)
Procedure
- Protein expression and purification
- Transform the pET15b vectors carrying Drp1 to Rosetta 2(DE3) pLysS competent cells.
- Plate the cells on an LB plate containing ampicillin and chloramphenicol (see Recipes) and incubate overnight at 37 °C.
- Select and place a colony into a 10 ml LB plate supplemented with ampicillin and chloramphenicol (see Recipes). Shake the bacteria culture overnight at 37 °C at 200 rpm (New Brunswick Scientific, series 25).
- Dilute the 5 ml of the culture into 1 L of the same growth medium and continue to grow the culture for 3-5 h at 37 °C using an incubator shaker at 200 rpm (New Brunswick Scientific, series 25).
- Cool down the cultured cells on ice, and add 0.2 ml of 0.5 M IPTG (see Recipes) to 1 L of LB buffer. Final IPTG concentration is 0.1 mM.
- Shake the culture overnight at 16 °C using a refrigerated incubator shaker (The Lab Companion, IS-971R) at 200 rpm with cooling system.
- After incubation, move the culture medium to a 1,000 ml bottle (Beckman Coulter) and centrifuge at 5,000 x g for 15 min at 4 °C in a JLA8.1000 rotor (Avanti J-26XP).
- Wash the bacteria twice with 40 ml PBS via centrifugation at 2,400 x g in a GH3.8 rotor using a CS-6R centrifuge (Beckman Coulter) for 15 min and freeze cell pellets at -80 °C.
- Resuspend the frozen cell pellets in 40 ml of lysis buffer (see Recipes).
- Sonicate the bacteria on ice for 10 x 5 sec at setting 5 and then 10 x 5 sec at setting 2 using a Fisher Scientific Sonic dismembrator model 100.
- Centrifuge the cell homogenate at 2,400 x g for 15 min at 4 °C in a GH3.8 rotor using a CS-6R centrifuge.
- Centrifuge the supernatant at 20,100 x g for 15 min at 4 °C in JA-20X (Avanti J-E).
- Pass the supernatant through a Millex-HA syringe filter with a pore size of 0.45 µm.
- Incubate the lysate with 2 ml of pre-washed 50% Ni-NTA beads (His-Bind Resin, Novagen) overnight at 4 °C using a Labquake Shaker Rotisserie.
- Load the sample in a 15-ml column (Poly-prep chromatography column, Bio-Rad Laboratories) in a cold room.
- Wash the beads with 3 ml of lysis buffer and then 15 ml of lysis buffer.
- Wash the beads with 3 ml of wash buffer (see Recipes) and then 15 ml of wash buffer.
- Elute His6-tagged recombinant Drp1 proteins from the column using 0.9 ml of elution buffer (see Recipes) three times and collect eluents into nine 300-µl fractions.
- Identify fractions containing Drp1 proteins using SDS-PAGE and Coomassie Brilliant Blue staining. Combine peak fractions (typically fractions #3 and 4) (Figure 1) and mix them with 15 ml of lysis buffer without imidazole.
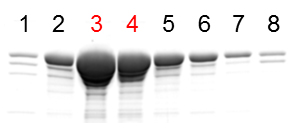
Figure 1. Analysis of purified Drp1 by SDS-PAGE and Coomassie Brilliant Blue staining - Pass the samples through an Amicon Ultra Centrifugation Filter (50k filter for full length Drp1 and 10k filter for domains) three times using lysis buffer without imidazole. The final volume is typically 1 ml.
- Mix the sample with final 20% DMSO, aliquoted, snap frozen in liquid nitrogen, and store at -80 °C. Typically concentrations of full-length Drp1 were 30 µM.
- Transform the pET15b vectors carrying Drp1 to Rosetta 2(DE3) pLysS competent cells.
- Flotation assay
- Unless otherwise noted, mix the lipids in a POPC:rhodamine-DPPE:variable lipid ratio of 84:1:15 (% mol).
- Dry up the lipids using nitrogen gas for 5 min and additionally dried in a SpeedVac overnight.
- Add 100 µl/one sample of 20 mM MES (pH 7.0)/100 mM NaCl (see Recipes) in lipid film and vortex for 1 h.
- Conduct five freeze-thaw cycles using dry ice and a 42 °C heat block.
- Generate the unilamellar liposomes via extrusion through a nanopore membrane with a pore size of 400 nm. This process is repeated 21 times.
- Confirm that the liposome is formed using fluorescence microscopy (Figure 2).
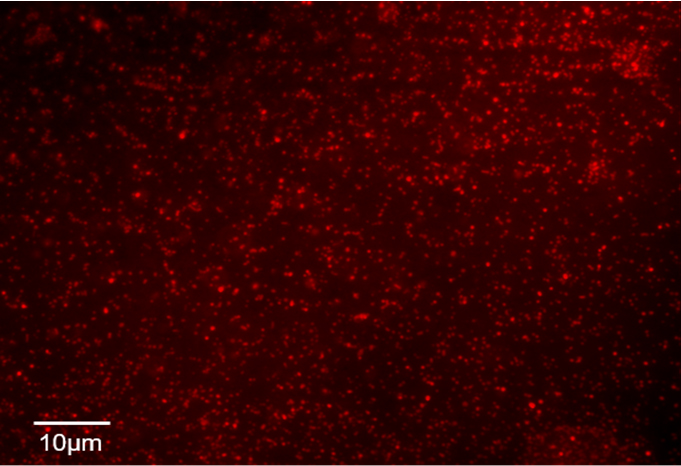
Figure 2. Confocal fluorescence microscopy of liposomes - Check the fluorescence intensity of rhodamine-PE by POLARstar Omega (Wavelength; Ex 544 nm, Em 590 nm) and then adjust the concentration of lipids based on the rhodamine intensity using 20 mM MES (pH 7.0)/100 mM NaCl.
- Mix the liposomes (5 mM lipids) with His6-Drp1 (5 μM) (final volume: 200 μl) and incubate at 4 °C with gentle mixing using a Labquake Shaker Rotisserie for 1 h in 20 mM MES (pH 7.0)/100 mM NaCl (Figure 3).
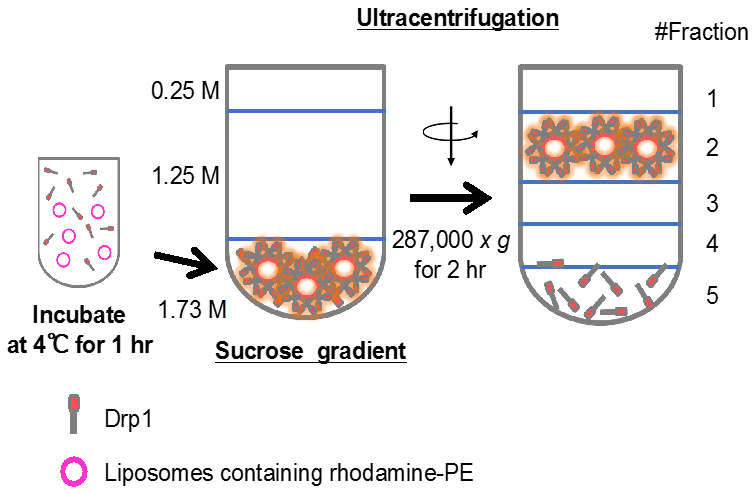
Figure 3. Drp1 and liposomes are incubated and analyzed by sucrose gradient centrifugation. Amounts of Drp1 that co-migrate with liposomes into fractions 1 and 2 are quantified. - Dilute the Drp1-liposome mixture in final 1.73 M sucrose/20 mM MES (pH 7.0) (final volume: 1.25 ml) placed at the bottom of tubes. Then overlay 2.9 ml of 1.25 M sucrose/20 mM MES (pH 7.0) and 0.85 ml of 0.25 M sucrose/20 mM MES (pH 7.0) (Figure 3).
- Centrifuge the sucrose gradient at 287,000 x g for 2 h at 4 °C in an SW55Ti rotor (Beckman Coulter).
- Collect the five fractions (0.5, 0.75, 1.75, 1.75 and 0.25 ml) from the top.
- The majority of the liposomes typically floats to the top two fractions.
- Measure the rhodamine intensity using a POLARstar Omega. Analyze each fraction (equal volume) using SDS-PAGE and silver staining (SilverQuestTM silver staining kit) according to the manufacturer’s instructions.
- Unless otherwise noted, mix the lipids in a POPC:rhodamine-DPPE:variable lipid ratio of 84:1:15 (% mol).
Data analysis
- Scan the stained gels using Scanmaker 8700, and quantify the band intensity using NIH ImageJ. Detect the liposomes based on the fluorescence intensity of rhodamine-PE. Normalize the amounts of Drp1 and liposomes relative to the volume of each fraction.
- For statistical analysis, P values are calculated using the Student’s t-test.
Notes
When loading the sample on SDS-page, please warm the sample with a heat block at 37 °C. Because the sample contains sucrose, the fluidity of the sample changes depending on the temperature.
Recipes
- LB plate containing ampicillin and chloramphenicol
10 g BactoTM tryptone
10 g NaCl
5 g BactoTM yeast extract
15 g agar
Fill up to 1 L with ultrapure water and autoclave at 121 °C for 30 min. After autoclaving, allow the agar solution to cool to 55 °C. Add final 100 µg/ml ampicillin and 25 µg/ml chloramphenicol - LB with ampicillin and chloramphenicol
10 g BactoTM tryptone
10 g NaCl
5 g BactoTM yeast extract
Fill up to 1 L with ultrapure water and autoclave at 121 °C for 30 min. After autoclaving, allow the solution to cool to 55 °C. Add final 100 µg/ml ampicillin and 25 µg/ml chloramphenicol - 0.5 M IPTG
1.19 g IPTG
Fill up to 10 ml with ultrapure water - 0.5 M HEPES (pH 7.4)
238.4 g/L HEPES
Adjust pH to 7.4 with KOH and fill up to 1 L with ultrapure water - Lysis buffer (10 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM 2-mercaptoethanol, 20 mM HEPES, pH 7.4)
40 ml/L 0.5 M HEPES (pH 7.4)
500 ml/L 1 M NaCl
1 ml/L 1 M MgCl2
5 ml/L 2 M imidazole
140 µl/L 2-mercaptoethanol
Fill up to 1 L with ultrapure water - Wash buffer (40 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM 2-mercaptoethanol, 20 mM HEPES, pH 7.4)
40 ml/L 0.5 M HEPES (pH 7.4)
500 ml/L 1 M NaCl
1 ml/L 1 M MgCl2
20 ml/L 2 M imidazole
140 µl/L 2-mercaptoethanol
Fill up to 1 L with ultrapure water - Elution buffer (250 mM imidazole, 1 mM MgCl2, 500 mM NaCl, 2 mM 2-mercaptoethanol, 20 mM HEPES, pH 7.4)
40 ml/L 0.5 M HEPES (pH 7.4)
500 ml/L 1 M NaCl
1 ml/L 1 M MgCl2
50 ml/L 2 M imidazole
140 µl/L 2-mercaptoethanol
Fill up to 1 L with ultrapure water - 10x PBS (pH 7.4)
2.56 g/L NaH2PO4·H2O
22.5 g/L Na2HPO4·7H2O
87.66 g/L NaCl
Fill up to 1 L with ultrapure water. Adjust pH to 7.0 with NaOH - Coomassie Brilliant Blue solution
2.5 g/L Coomassie Brilliant Blue R-250
100 ml/L acetic acid
450 ml/L MeOH
450 ml/L ultrapure water - 100 mM MES (pH 7.0)
21.538 g/L MES
3.068 g/L NaOH
Fill up to 1 L with ultrapure water - 20 mM MES (pH 7.0)
200 ml/L 100 mM MES (pH 7.0)
Fill up to 1 L with ultrapure water. Adjust pH to 7.0 with NaOH - 20 mM MES (pH 7.0)/100 mM NaCl
200 ml/L 100 mM MES (pH 7.0)
5.844 g/L NaCl
140 µl/L 2-mercaptoethanol (final 2 mM)
Fill up to 1 L with ultrapure water. Adjust pH to 7.0 with NaOH, as necessary - 2.7 M sucrose/20 mM MES/100 mM NaCl
924.21 g/L sucrose
Fill up to 1 L with 20 mM MES/100 mM NaCl - 1.25 M sucrose/20 mM MES/100 mM NaCl
427.875 g/L sucrose
Fill up to 1 L with 20 mM MES/100 mM NaCl - 0.25 M sucrose/20 mM MES/100 mM NaCl
85.575 g/L sucrose
Fill up to 1 L with 20 mM MES/100 mM NaCl
Acknowledgments
We thank past and present members of the Iijima and Sesaki labs for helpful discussions and technical assistance. This work was supported by NIH grants to M.I. (GM084015) and H.S. (GM089853).
References
- Adachi, Y., Itoh, K., Yamada, T., Cerveny, K. L., Suzuki, T. L., Macdonald, P., Frohman, M. A., Ramachandran, R., Iijima, M. and Sesaki, H. (2016). Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol Cell 63(6): 1034-1043.
- Bustillo-Zabalbeitia, I., Montessuit, S., Raemy, E., Basanez, G., Terrones, O. and Martinou, J. C. (2014). Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One 9(7): e102738.
- Macdonald, P. J., Stepanyants, N., Mehrotra, N., Mears, J. A., Qi, X., Sesaki, H. and Ramachandran, R. (2014). A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell 25(12): 1905-1915.
- Montessuit, S., Somasekharan, S. P., Terrones, O., Lucken-Ardjomande, S., Herzig, S., Schwarzenbacher, R., Manstein, D. J., Bossy-Wetzel, E., Basanez, G., Meda, P. and Martinou, J. C. (2010). Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142(6): 889-901.
- Roy, M., Reddy, P. H., Iijima, M. and Sesaki, H. (2015). Mitochondrial division and fusion in metabolism. Curr Opin Cell Biol 33: 111-118.
- Stepanyants, N., Macdonald, P. J., Francy, C. A., Mears, J. A., Qi, X. and Ramachandran, R. (2015). Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell 26(17): 3104-3116.
- Tamura, Y., Itoh, K. and Sesaki, H. (2011). SnapShot: Mitochondrial dynamics. Cell 145(7): 1158, 1158 e1151.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Adachi, Y., Itoh, K., Iijima, M. and Sesaki, H. (2017). Assay to Measure Interactions between Purified Drp1 and Synthetic Liposomes. Bio-protocol 7(9): e2266. DOI: 10.21769/BioProtoc.2266.
Category
Biochemistry > Lipid > Lipid-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









