- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analyses of Root-secreted Acid Phosphatase Activity in Arabidopsis
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2202 Views: 12838
Reviewed by: Marisa RosaNing LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1812 Views
Abstract
Induction and secretion of acid phosphatase (APase) is a universal adaptive response of higher plants to low-phosphate stress (Tran et al., 2010). The intracellular APases are likely involved in the remobilization and recycling of phosphate (Pi) from intracellular Pi reserves, whereas the extracellular or secreted APases are believed to release Pi from organophosphate compounds in the rhizosphere. The phosphate starvation-induced secreted APases can be released into the rhizosphere or retained on root surfaces (root-associated APases). In this article, we describe the protocols for analyzing root-secreted APase activity in the model plant Arabidopsis thaliana (Arabidopsis). In Arabidopsis, the activity of both root-associated APases and APases that are released into the rhizosphere can be quantified based on their ability to cleave a synthesized substrate, para-nitrophenyl-phosphate (pNPP), which releases a yellow product, para-nitrophenol (pNP) (Wang et al., 2011 and 2104). The root-associated APase activity can also be directly visualized by applying a chromogenic substrate, 5-bromo-4-chloro-3-indolyl-phosphate (BCIP), to the root surface (Lloyd et al., 2001; Tomscha et al., 2004; Wang et al., 2011 and 2014) whereas the isozymes of APases that are released into rhizosphere can be profiled using an in-gel assay (Trull and Deikman, 1998; Tomscha et al., 2004; Wang et al., 2011 and 2014). The protocol for analysis of intracellular APase activity in Arabidopsis has been previously described (Vicki and William, 2013).
Keywords: Arabidopsis thalianaBackground
Phosphate (Pi) is the major form of phosphorus that plants take up through their root systems, and Pi levels in most soils are low, resulting in Pi starvation. To cope with this nutritional stress, plants trigger an array of adaptive responses that increase their survival and growth. Induction and secretion of APases is a hallmark Pi starvation response that has been documented in a variety of plant species (Tran et al., 2010), and root-secreted APase activity has been widely used as a diagnostic tool to evaluate the magnitude of plant responses to Pi starvation. In this article, we provide three protocols for assay of root-secreted APase activity. An assay using pNPP as a substrate is the most commonly used method to quantify APase activity. A BCIP staining assay provides a simple, one-step method for the histochemical detection of APase activity on the root surface. Neither of these methods, however, can reveal the number of APase isoforms that contribute to the observed activity. A third method (in-gel assay), which combines electrophoresis, protein renaturation, and conversion of a substrate into a red-brown product, can reveal the diverse compositions of APase isoforms in different samples, and can therefore provide additional information about root-secreted APases.
Materials and Reagents
- 9-cm-diameter Petri dish
- Surgical blade (Swann Morton Surgical Scalpel Blade No. 20)
- 1.7 ml and 2 ml Eppendorf tubes
- Dialysis tubing (Sigma-Aldrich, catalog number: D9777 )
- 50 ml Falcon tube
- Whatman #1 filter paper (GE Healthcare, catalog number: 1001-125 )
- Pipette tips
- Arabidopsis seeds
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- 30% acrylamide/Bis solution (Solarbio, catalog number: A1010 )
- TEMED (Sigma-Aldrich, catalog number: T22500 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Bovine serum albumin (BSA)*
- 95% ethanol*
- Ammonium persulfate (NH4)2S2O8*
- Sodium hypochlorite solution (NaClO)*
- Sodium hydroxide (NaOH)*
- Sodium acetate (NaAc)*
- Phosphoric acid (H3PO4)*
- Hydrochloric acid (HCl)*
- Ammonium nitrate (NH4NO3)*
- Potassium nitrate (KNO3)*
- Calcium chloride (CaCl2)*
- Magnesium sulfate (MgSO4)*
- Potassium dihydrogen phosphate (KH2PO4)*
- Potassium sulfate (K2SO4)*
- Potassiu iodide (KI)*
- Orthoboric acid (H3BO3)*
- Manganese sulfate (MnSO4)*
- Zinc sulfate (ZnSO4)*
- Sodium molybdate (Na2MoO4)*
- Cobalt(II) chloride (CoCl2)*
- Copper sulfate (CuSO4)*
- Fe-EDTA*
- Myo Inositol*
- Nicotinic acid*
- Pyridoxine HCl*
- Thiamine HCl*
- Glycine*
- Sucrose*
- Bromophenol blue*
- Coomassie Brilliant Blue G-250*
- MES (AMRESCO, catalog number: E169 )
- β-naphthyl acid phosphate (Sigma-Aldrich, catalog number: N7375 )
- Fast Black K (Sigma-Aldrich, catalog number: F7253 )
- pNPP (Sigma-Aldrich, catalog number: P4744 )
- BCIP (Gold Bio, catalog number: B-500-10 )
- Tris base (NOVON SCIENTIFIC, catalog number: ZZ02531 )
- DTT (Sigma-Aldrich, catalog number: DTT-RO )
- SDS (AMRESCO, catalog number: 0227 )
- DMSO (AMRESCO, catalog number: 0231 )
- PMSF (Sigma-Aldrich, catalog number: 52332 )
- Liquid nitrogen
- Murashige and Skoog (MS) medium (P+ and P- MS medium) (see Recipes, Murashige and Skoog, 1962)
- APase activity profiling detection buffer, pH 4.9 (see Recipes)
- Reaction buffer, pH 4.9 (see Recipes)
- pNPP solution (1.0 mg/ml) (see Recipes)
- BCIP stock solution (100x) (see Recipes)
- Bradford solution (see Recipes)
- 5x protein loading buffer (see Recipes)
*Note: Similar reagents from any qualified company are suitable for this experiment.
Equipment
- Pipettes
- Forceps (Fisher Scientific, catalog number: 22-327379 )
- Spectrophotometer (WPA, Biowave)
- 250 ml Erlenmeyer (conical) flask
- Orbital shaker (Qilinbeier, model: TS-1 )
- Freeze dryer (Beijing Songyuanhuaxing, model: LGJ-10 )
- Microfuge (Eppendorf, model: 5415 D )
- Microwave oven
- Small mortar and pestle
- Water bath (Shanghai Yiheng, model: DK-8D )**
- Analytical balance (Mettler Toledo, model: PB153-L)**
- pH meter (Mettler Toledo, model: FE20 )**
- Mini-PROTEAN® Tetra Vertical Electrophoresis Cell (Bio-Rad Laboratories, model: Mini-PROTEAN® Tetra Vertical Electrophoresis Cell , catalog number: 1658004)
**Note: Similar equipment from any qualified company is suitable for this experiment.
Software
- SPSS Statistics
Procedure
- Quantification of root-associated APase activity and the activity of APases released into the growth media
- Quantification of root-associated APase activity
- Arabidopsis seeds are suspended with 5% sodium hypochlorite solution/0.1% Triton X-100 and incubated for 10 min at room temperature with occasionally inverting for surface-sterilization.
- Remove the liquid and wash the seeds with sterilized ddH2O for 4 times.
- Place 20-30 surface-sterilized Arabidopsis seeds horizontally on the upper part of each 9-cm-diameter Petri dish containing full-strength or half-strength Murashige and Skoog (MS) medium (P+ or P- medium). For each biological sample, use three replicates.
- Keep the Petri dishes at 4 °C for 2 days for stratification.
- Place the Petri dishes vertically in a controlled environment growth room and allow the seedlings to grow for 6-7 days under a 16-h light/8-h dark regime (90 µmol photons m-2 sec-1) at 22-23 °C.
- Excise roots with a razor blade and measure root length. The average root length of the Arabidopsis seedlings grown on P+ medium is 4.0-5.5 cm and that grown on P- medium is 2.5-4.0 cm.
- Immerse two roots with forceps into 650 µl of pre-warmed (37 °C) reaction buffer in a 1.7 ml Eppendorf tube.
- Quickly add 50 µl of pNPP solution.
- Incubate at 37 °C for 1-4 h.
- Add 100 µl of 0.4 N NaOH and shake vigorously by hand to stop the reaction.
- Remove the roots with forceps and measure the absorbance at 410 nm (A410) in the remaining solution with a spectrophotometer.
- Calculate the root-associated APase activity as A410/cm root/h (Figure 1A).
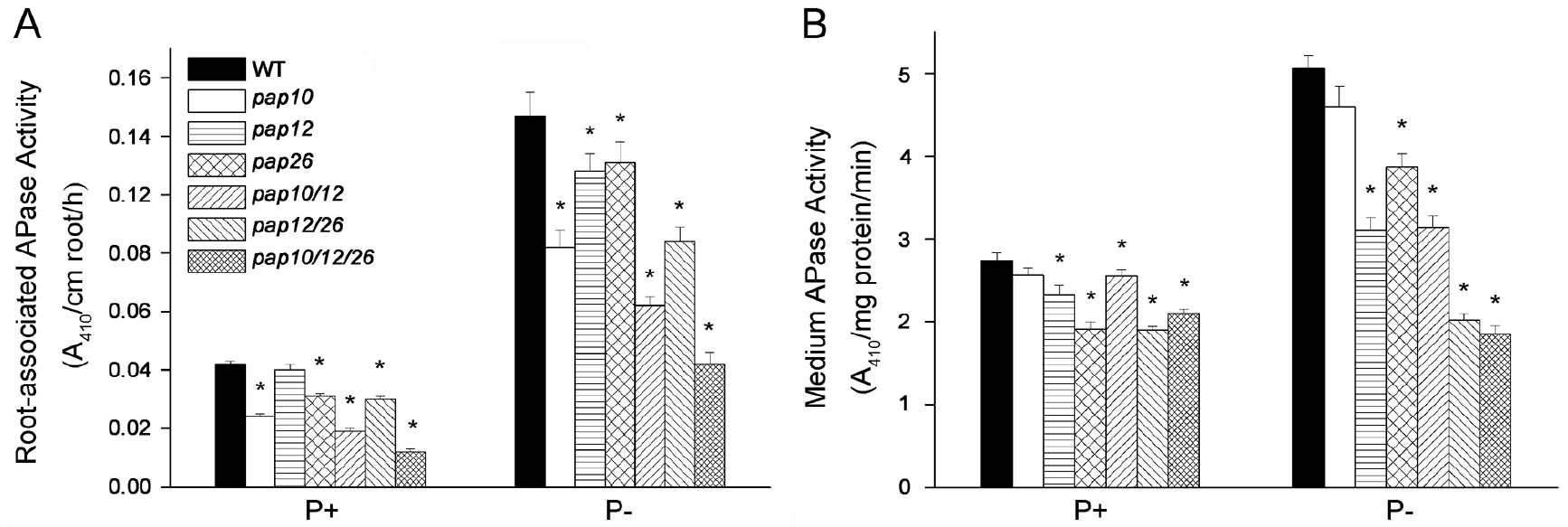
Figure 1. Analysis of APase activity secreted by Arabidopsis seedlings of WT and various single, double, and triple pap (purple acid phosphatase) mutants grown in or on P+ or P- medium. Root-associated APase activities (A) and APase activities in the medium (B) were determined with pNPP as the substrate. Values are means with SE. A one-way ANOVA analysis was carried out for the whole data set, and post hoc comparisons were conducted using the SPSS Tukey HSD test at P < 0.05. Within each of the four plots, values with different letters are significantly different. Adapted from Wang et al. (2014).
- Arabidopsis seeds are suspended with 5% sodium hypochlorite solution/0.1% Triton X-100 and incubated for 10 min at room temperature with occasionally inverting for surface-sterilization.
- Concentrating the proteins in the liquid growth medium
- Transfer about 150 9-day-old seedlings that were grown vertically on P+ solid medium with autoclaved forceps to a 250 ml autoclaved conical flask containing 100 ml of P+ or P- liquid medium.
- Place the flasks on an orbital shaker (40 rpm/min) and allow the seedlings to grow for 6-7 days under a 16-h light/8-h dark regime (90 µmol photons m-2 sec-1) at 22-23 °C.
- Remove all of the seedlings from the flasks and freeze the liquid medium at -20 °C.
- Put the medium in the holder of a pre-cooled freeze dryer, cover the lid, and lyophilize the medium for 3-4 days.
- Thoroughly wash the bottom of the flask using 5-6 ml of ddH2O and transfer the washing solution to dialysis tubing.
- Dialyze the sample for 24 h at 4 °C against 4 L of pre-cooled ddH2O and change the ddH2O every 6 h.
- Transfer the dialyzed sample to a 2 ml tube and centrifuge for 5 min at 1,500 x g.
- Transfer the supernatant to a 50 ml Falcon tube and freeze the liquid at -20 °C, then lyophilize for 2 days as described in step A2d.
- Dissolve the lyophilized sample in 200 µl of ddH2O.
- Measure the protein concentration using the Bradford method as described in step A3.
- The concentrated, secreted proteins can be used immediately or can be stored at -80 °C for further analysis.
- Transfer about 150 9-day-old seedlings that were grown vertically on P+ solid medium with autoclaved forceps to a 250 ml autoclaved conical flask containing 100 ml of P+ or P- liquid medium.
- Determination of protein concentration
- Pipette 0, 2, 4, 8, 12, 16, or 20 μl of 1 μg/μl BSA into 1.7 ml Eppendorf tubes and add ddH2O to a final volume of 20 μl.
- Add 980 μl of Bradford solution to each tube and mix well.
- Incubate at room temperature for 5 min.
- Measure the absorbance at 595 nm (A595) with a spectrophotometer.
- Make a Bradford standard curve by plotting BSA content and A595.
- Add 20 μl of concentrated secreted proteins to 980 μl of Bradford solution.
- Mix well and incubate at room temperature for 5 min.
- Measure the A595 with a spectrophotometer.
- Compare the resulting A595 value with the Bradford standard curve to determine the protein concentration of various samples.
- Pipette 0, 2, 4, 8, 12, 16, or 20 μl of 1 μg/μl BSA into 1.7 ml Eppendorf tubes and add ddH2O to a final volume of 20 μl.
- Quantification of APase activity in the growth medium
- Add 10 µl of concentrated proteins (about 2.5 µg) collected from the liquid growth medium to 590 µl of reaction buffer in a 1.7 ml Eppendorf tube. For each sample, make a least three replicates.
- Add 80 μl of pNPP solution to the reaction mixture.
- Incubate at 37 °C for 1 h.
- Add 120 μl of 0.4 N NaOH to terminate the reaction.
- Measure the A410 with a spectrophotometer.
- Calculate the APase activity in the growth medium as A410/mg protein/min (Figure 1B).
- Add 10 µl of concentrated proteins (about 2.5 µg) collected from the liquid growth medium to 590 µl of reaction buffer in a 1.7 ml Eppendorf tube. For each sample, make a least three replicates.
- Quantification of root-associated APase activity
- Analysis of root-associated APase activity using histochemical staining
- Grow seedlings vertically on P+ or P- MS medium for 6-7 days as described in steps A1a-A1e.
- Add 100 ml of ddH2O and 0.5 g agar to a 250 ml conical flask, gently shake the flask, and then heat the flask in a microwave oven until the agar is completely dissolved.
- Add 1 ml of the BCIP stock solution (100x) and shake briefly by hand.
- Keep the BCIP/agar solution at room temperature or use flowing water to cool the solution for a few minutes to about 40 °C.
- Keep the BCIP/agar solution in a 40 °C water bath.
- Uniformly overlay about 5 ml of the BCIP/agar solution on one row of the Arabidopsis roots and incubate at room temperature for about 24 h (Figure 2).
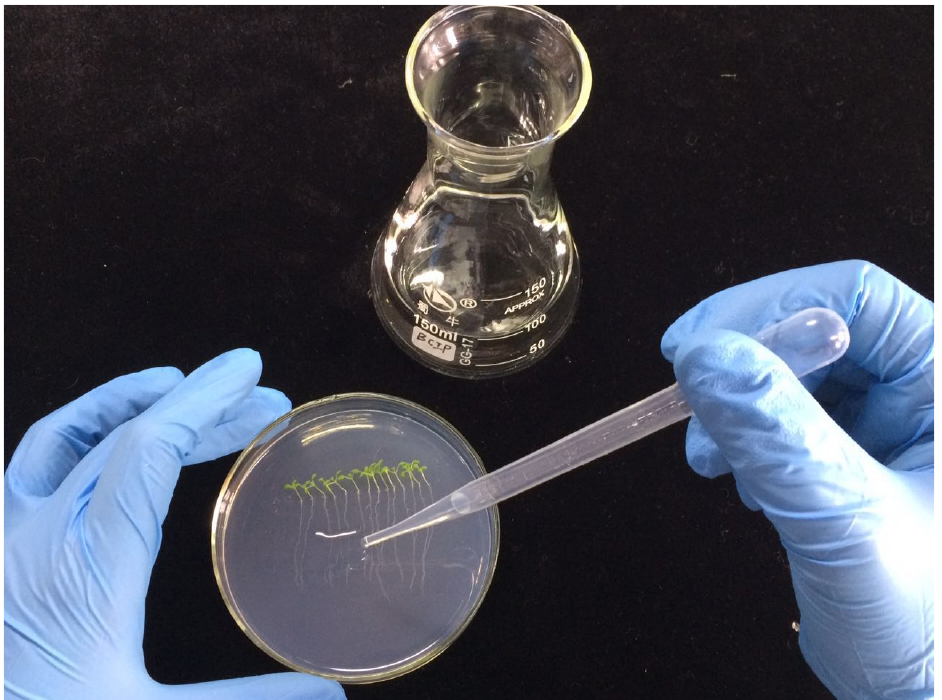
Figure 2. A demonstration of how to overlay the BCIP/agar solution on the root surface of Arabidopsis seedlings grown on Petri dish - The roots of Arabidopsis grown on P- medium will be stained blue, while these grown on P+ medium will remain unstained (Figure 3).
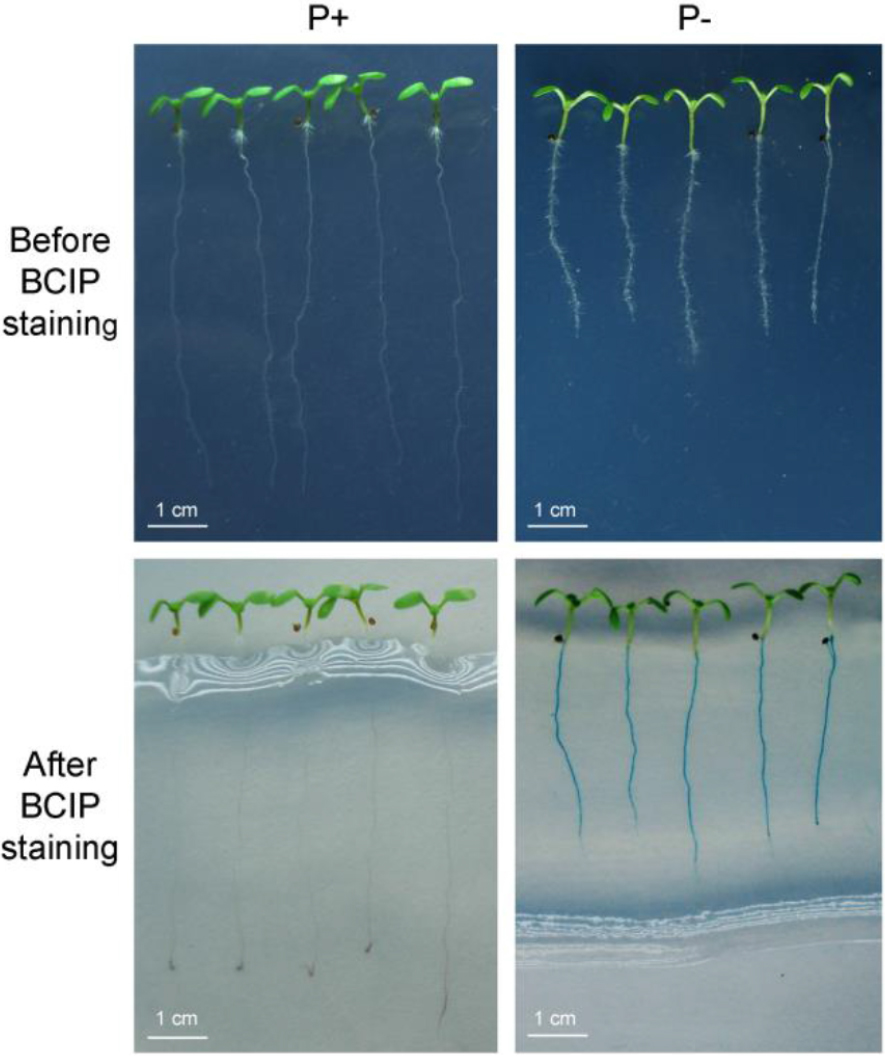
Figure 3. Histochemical staining of root-associated APase activity. Upper row: The morphologies of 7-day-old Arabidopsis seedlings grown on MS P+ and P- medium without BCIP staining. Bottom row: Seedlings whose roots were stained with a 0.01% BCIP/0.5% agar solution for 24 h. In the lower row, root-associated APase activity (as indicated by the blue color) was detected on roots of seedlings grown on P- medium (bottom right) but not on roots of seedlings grown on P+ medium (bottom left). - APase histochemical staining is greatly affected by the BCIP concentration. A high concentration of BCIP (0.08%-0.16%) can cause over-staining, which may mask differences in the level of secreted APase activity (Figure 4).
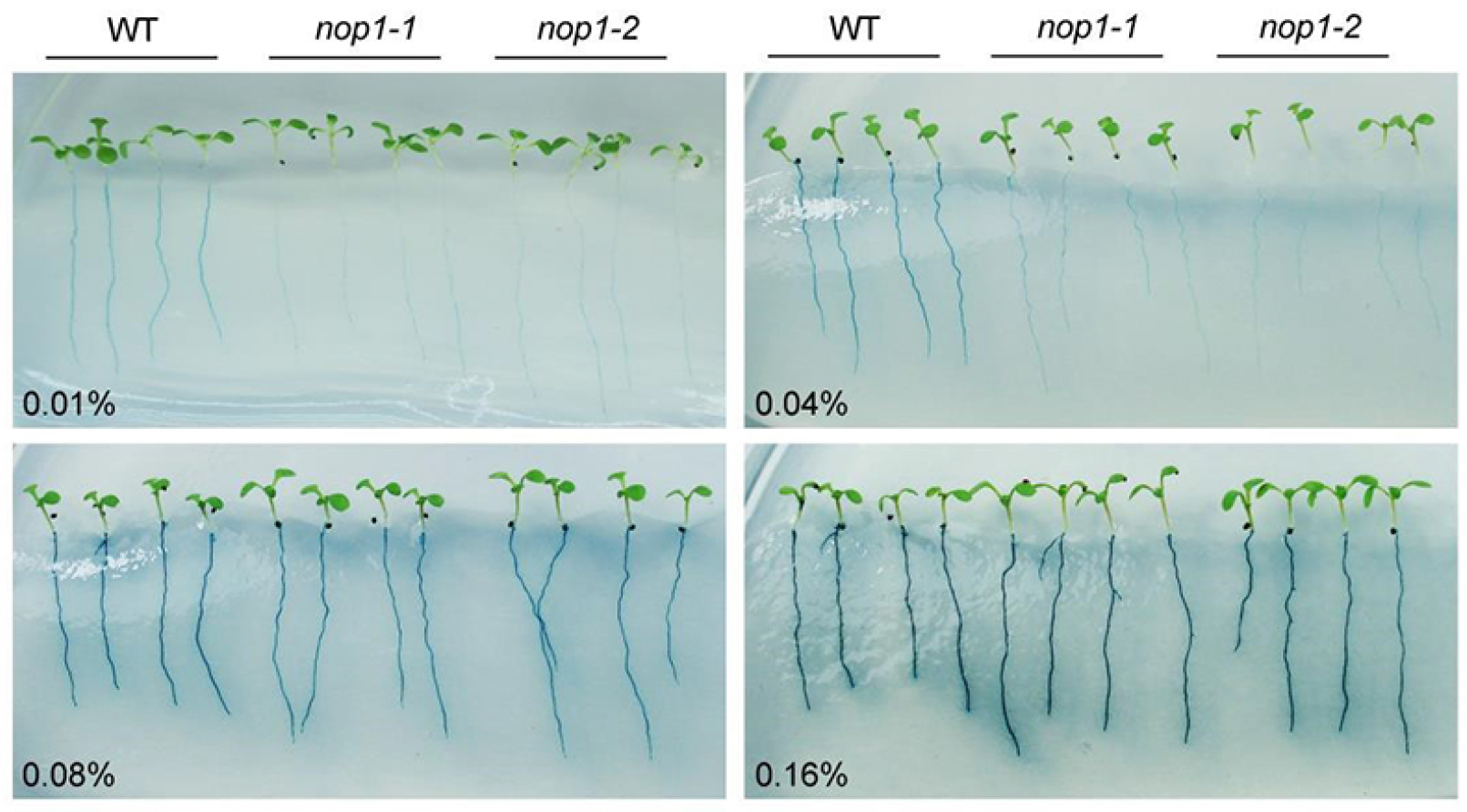
Figure 4. The effect of different concentrations of BCIP on staining of root-associated APase activity of 7-day-old Arabidopsis seedlings grown on MS P- medium. The concentrations of BCIP used are indicated at the left bottom corner. nop1-1 and nop1-2 are Arabidopsis mutants that have mutations in the AtPAP10 gene. With the increase in BCIP concentration, the activities of other root-associated APases, mainly AtPAP12 and AtPAP26, are also detected. Adapted from Wang et al. (2011).
- Grow seedlings vertically on P+ or P- MS medium for 6-7 days as described in steps A1a-A1e.
- Analysis of profiles of APase isoforms released into the growth medium (in-gel assay)
The isozyme profile of APases released into the growth medium is visualized by separating protein samples on SDS-PAGE (denaturing gel) or Native-PAGE (non-denaturing gel), which are then stained with the chromogenic substrates β-NAP and fast black K. For Native-PAGE analysis, omit SDS from the gel and running buffer.
Prepare a 10% separating gel and a 5% stacking gel (SDS-PAGE) according to the following Table 1.
Table 1. Recipes for separating gel and stacking gelStock solution5% stacking gel (5 ml)10% separating gel (10 ml)30% Acrylamide/Bis solution0.83 ml3.3 ml1.5 M Tris-HCl, pH 8.8-2.5 ml1 M Tris-HCl, pH 6.80.63 ml-10% SDS0.05 ml0.1 ml10% (NH4)2S2O80.05 ml0.1 mlTEMED0.005 ml0.004 mlddH2O3.40 ml4.0 ml- Set up the SDS-PAGE gel running system according to the Bio-Rad instructions (http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6040.pdf).
- Add 1x protein running buffer that was pre-cooled to 4 °C.
- Thoroughly mix 2.5 μg of secreted proteins with 1/5 volume of 5x protein loading buffer and load the sample on the SDS-PAGE gel.
Note: Do not heat the sample. - Separate proteins at 60 V for 30 min followed by 120 V for 150 min (both at 4 °C).
- Carefully transfer the gel to pre-cooled (4 °C) ddH2O in a container and gently shake for 10 min to remove SDS from the gel.
- Change pre-cooled (4 °C) ddH2O and repeat 4 times.
- Add 50 ml of reaction buffer and shake gently for 15 min at 4 °C.
- Change reaction buffer and repeat 4 times.
- Add 50 ml of APase activity profiling detection buffer and incubate at 37 °C for 1-3 h with gentle agitation. The isozyme profile of APases is visualized as the red-brown bands on the gel (Figure 5).
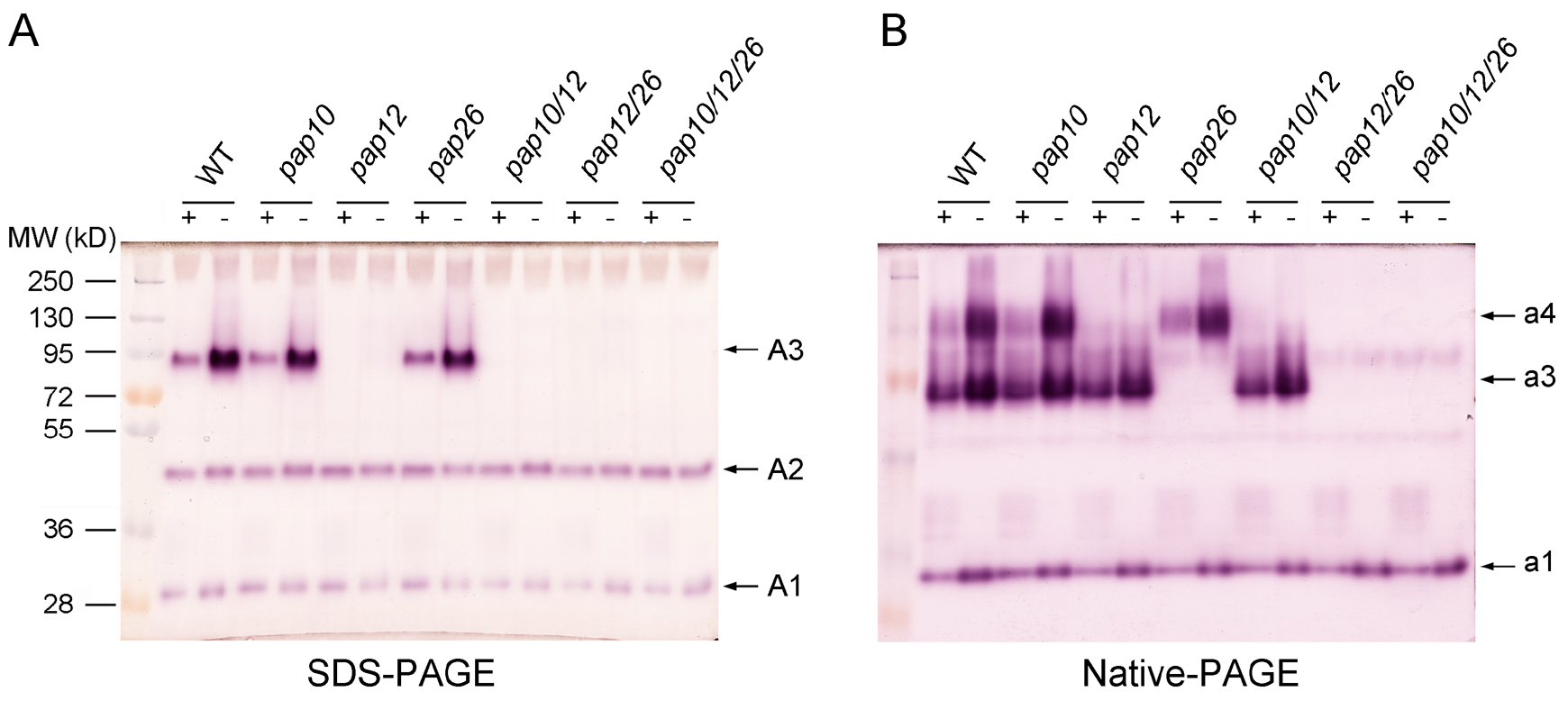
Figure 5. Profiles of APase activity in the growth medium. A 2.5-μg quantity of secreted protein was separated by SDS-PAGE (A) or Native-PAGE (B) and stained with 0.3 mg/ml β-NAP and 0.5 mg/ml Fast Black K. Protein molecular markers are on the left side of the gel in (A). The numbers on the right of each panel indicate different APase isoforms. Adapted from Wang et al. (2014).
- Set up the SDS-PAGE gel running system according to the Bio-Rad instructions (http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6040.pdf).
Notes
- The temperature of BCIP/agar solution is important for detecting root-associated APase activity. If the solution is too hot (above 60 °C), it will damage Arabidopsis roots and inactivate the APase on the root surface.
- Removal of SDS is critical for obtaining a clear profile of APase isozymes.
Recipes
- Murashige and Skoog (MS) medium (P+ and P- MS medium)
P+ MS medium is the full-strength MS medium (Murashige and Skoog, 1962). The recipe for full-strength MS medium is shown below. For P- MS medium, the 1.25 mM KH2PO4 in P+ MS medium is replaced with 1.25 mM K2SO4. Autoclave P+ and P- medium at 121 °C for 10 min, and store the medium at room temperatureNH4NO3 20.62 mM CuSO4 0.1 µM KNO3 18.79 mM Fe-EDTA 100 µM CaCl2 2.99 mM Myo inositol 0.55 µM MgSO4 1.5 mM Nicotinic acid 4.1 µM KH2PO4 1.25 mM Pyridoxine HCl 2.4 µM K2SO4 0.625 mM Thiamine HCl 0.3 µM KI 5 µM Glycine 30 µM H3BO3 100 µM MnSO4 92.5 µM Sucrose 10 g/L ZnSO4 30 µM MES 5.1 g/L NaMoO4 1 µM pH 5.8 CoCl2 0.1 µM Agar 10 g/L - APase activity profiling detection buffer, pH 4.9 (freshly prepared)
0.3 mg/ml β-naphtyl acid phosphate
0.5 mg/ml Fast Black K
10 mM MgCl2
50 mM NaAc - Reaction buffer, pH 4.9
10 mM MgCl2
50 mM NaAc
Store at 4 °C - pNPP solution (1.0 mg/ml)
A 10-mg quantity of pNPP is dissolved in 10 ml of reaction buffer
Store aliquots at -20 °C - BCIP stock solution (100x)
The concentration of the BCIP stock solution is 10 g/L, and that of the working solution is 0.1 g/L
To prepare the BCIP stock solution: - Dissolve 0.1 g of BCIP powder in 8 ml of ddH2O and then add approximately 14 drops of 1 N NaOH until the BCIP power is totally dissolved
- Add ddH2O to a total volume of 10 ml and then filter the solution to remove un-dissolved BCIP particles
- Store aliquots at -20 °C
- Bradford solution
Dissolve 100 mg of Coomassie Brilliant Blue G-250 in 50 ml of 95% ethanol
Add 100 ml of 85% H3PO4 and then add ddH2O to 1,000 ml
Pass through a Whatman #1 filter paper, and store the filtrate at 4 °C in a brown bottle - 5x protein loading buffer
250 mM Tris-HCl, pH 6.8
10 mM DTT
10% SDS
30% (v/v) glycerol
0.05% (w/v) bromophenol blue
Store the buffer at room temperature
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant Nos. 31370290 and 30670170).
References
- Lloyd, J. C., Zakhleniuk, O. V. and Raines, C. A. (2001). Identification of mutants in phosphorus metabolism. Annals Applied Biol 138: 111-115.
- Murashige, T and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15(3): 473-497.
- Tomscha, J. L., Trull, M. C., Deikman, J., Lynch, J. P. and Guiltinan, M. J. (2004). Phosphatase under-producer mutants have altered phosphorus relations. Plant Physiol 135(1): 334-345.
- Tran, H. T., Hurley, B. A. and Plaxton, W. C. (2010). Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14-27.
- Trull, M. C. and Deikman, J. (1998). An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206(4): 544-550.
- Vicki, K. and William, P. (2013). Protein extraction, acid phosphatase activity assays, and determination of soluble protein concentration. Bio-protocol e889.
- Wang, L., Li, Z., Qian, W., Guo, W., Gao, X., Huang, L., Wang, H., Zhu, H., Wu, J. W., Wang, D. and Liu, D. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157(3): 1283-1299.
- Wang, L., Lu, S., Zhang, Y., Li, Z., Du, X. and Liu, D. (2014). Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56(3): 299-314.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Wang, L. and Liu, D. (2017). Analyses of Root-secreted Acid Phosphatase Activity in Arabidopsis. Bio-protocol 7(7): e2202. DOI: 10.21769/BioProtoc.2202.
- Wang, L., Li, Z., Qian, W., Guo, W., Gao, X., Huang, L., Wang, H., Zhu, H., Wu, J. W., Wang, D. and Liu, D. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157(3): 1283-1299.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










