- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
MHC Class II Tetramer Labeling of Human Primary CD4+ T Cells from HIV Infected Patients
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2187 Views: 11421
Reviewed by: Jia LiGuangzhi ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Multi-color Flow Cytometry Protocol to Characterize Myeloid Cells in Mouse Retina Research
Wei Xiao [...] Abdelrahman Y. Fouda
Aug 20, 2023 2980 Views

Isolation and Analysis of B-cell Progenitors from Bone Marrow by Flow Cytometry
Hongchang Zhao [...] Jacqueline H. Barlow
Oct 5, 2023 3475 Views

Addressing the Role of Conventional CD8αβ+ T Cells and CD4+ T Cells in Intestinal Immunopathology Using a Bone Marrow–Engrafted Model
Amneh Aoudi [...] Saïdi Soudja
Feb 20, 2024 2955 Views
Abstract
Major Histocompatibility Complex (MHC) tetramers have been used for two decades to detect, isolate and characterize T cells specific for various pathogens and tumor antigens. In the context of Human Immunodeficiency Virus (HIV) infection, antigen-specific CD8+ T cells have been extensively studied ex vivo, as they can be readily detected by HIV peptide-loaded MHC class I tetramers. In contrast, the detection of HIV-specific CD4+ T cells has proven more challenging, due to the intrinsically lower clonal expansion rates of CD4+ T cells, and to the preferential depletion of HIV-specific CD4+ T cells in the course of HIV infection.
In the following protocol, we describe a simple method that facilitates the identification of CD4+ T cells specific for an HIV-1 capsid epitope using peptide-loaded MHC class II tetramers. Tetramer labeled CD4+ T cells can be analyzed for their cell surface phenotype and/or FACS-sorted for further downstream applications. A key point for successful detection of specific CD4+ T cells ex vivo is the choice of a peptide/MHC II combination that results in high-affinity T Cell Receptor (TCR) binding (Benati et al., 2016). A second key point for reliable detection of MHC II tetramer-positive cells is the systematic use of a control tetramer loaded with an irrelevant peptide, with the sample and control tubes being processed in identical conditions.
Background
Rare HIV-specific MHC II tetramer-positive cells have been detected in purified CD4+ T cells, after magnetic enrichment of tetramer-PE labeled cells with anti-PE microbeads (Seth et al., 2005). We found that with validated peptide/MHC II tetramer combinations, a simpler protocol based on direct tetramer labeling of 5 x 106 patient Peripheral Blood Mononuclear Cells (PBMC), followed by acquisition of all events on a flow cytometer, resulted in reliable detection of HIV-specific CD4+ T cells. Exclusion of irrelevant cells (CD14+, CD20+, CD8+) and dead cells (Fixable Viability dye+) through an appropriate gating strategy improved labeling specificity.
Materials and Reagents
- Falcon® round-bottom 5 ml polypropylene tubes (Corning, Falcon®, catalog number: 352063 )
- Falcon® round-bottom 5 ml polystyrene tubes with a 35 µm Cell-Strainer cap (Corning, Falcon®, catalog number: 352235 )
- APC-conjugated antigen-loaded MHC II tetramers can be obtained through the NIH Tetramer Core Facility, Emory University, USA. The tetramers are provided at a concentration in the 1-1.5 mg/ml range, in aliquots of 200 µl
- APC-conjugated MHC II tetramers loaded with an irrelevant peptide (usually the CLIP peptide: PVSKMRMATPLLMQA can be obtained through the NIH Tetramer Core Facility, Emory University, USA)
- Mouse-anti-human CD3 APC-eFluor® 780 (clone UCHT1) (Affymetrix, eBioscience, catalog number: 47-0038-42 )
- Mouse-anti-human CD4 BD HorizonTM PE-CF594 (clone RPA-T4) (BD, BD Biosciences, catalog number: 562281 )
- Mouse-anti-human CD8 Brilliant Violet 785TM (clone RPA-T8) (BioLegend, catalog number: 301045 )
- Mouse-anti-human CD14 VioGreen® (clone TÜK4) (Miltenyi Biotec, catalog number: 130-096-875 )
- Mouse-anti-human CD20 VioGreen® (clone LT20) (Miltenyi Biotec, catalog number: 130-096-904 )
- Fixable Viability Dye eFluor 506® (Affymetrix, eBioscience, catalog number: 65-0866-14 )
- 16% paraformaldehyde (PFA) solution (Electron Microscopy Sciences, catalog number: 15710 )
- Phosphate buffered saline (PBS), pH 7.4 (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- RPMI 1604 GlutaMAX ITM (Thermo Fisher Scientific, GibcoTM, catalog number: 61870044 )
- HyCloneTM fetal bovine serum (FBS) (South America), research grade (GE Healthcare, catalog number: SV30160.03 )
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- 1 M HEPES buffer (Dominique Dutscher SAS, catalog number: P05-01100P )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912 )
- Sodium azide (NaN3) 5% (w/v) (VWR, BDH®, catalog number: BDH7465-2 )
- Human AB serum (PAN-Biotech, catalog number: P30-2901 )
- Complete RPMI (see Recipes)
- FACS buffer (see Recipes)
- Tetramer labeling buffer (see Recipes)
- FACS sorting buffer (see Recipes)
Equipment
- Benchtop centrifuge (Thermo Fisher Scientific, model: SorvallTM LegendTM XTR )
- Benchtop microcentrifuge (Eppendorf, model: 5254 R )
- BD LSRFortessaTM cell analyzer (BD, BD Biosciences, model: BD LSRFortessaTM Cell Analyzer )
- BD FACSAria IITM flow cytometer (BD, BD Biosciences, model: BD FACSAria IITM Cell Sorter )
Software
- FACSDivaTM version 8.0 (BD)
- FlowJoTM version 10.2 (FlowJo, LLC)
Procedure
Ex vivo tetramer labeling of CD4+ T cells from HIV infected patients
- PBMC from HIV infected samples are either thawed from frozen vials or freshly processed from whole blood in a biosafety level-3 environment.
- 15-50 x 106 cells are dispatched into 5 ml polypropylene tubes (5 x 106 cells per tube) and washed once with 2 ml complete RPMI at 600 x g for 7 min at 4 °C.
Notes:- A wash entails the addition of the indicated volume of the specified washing buffer followed by centrifugation and careful removal of the supernatant.
- A minimum of 5 x 106 PBMC is required per tetramer labeling; the same number of PBMC is required for the control tetramer labeling. When scaling up, it is preferable to process multiple identical sample tubes in parallel, so that the amount of cells required for the control tube remains constant at 5 x 106 PBMC.
- Individual tubes containing 0.5 x 106 cells in 50 μl FACS buffer should be prepared at this stage to be used as single stained compensation controls. These tubes should be kept at 4 °C for the duration of the tetramer labeling until the point of addition of cell surface antibodies.
- A wash entails the addition of the indicated volume of the specified washing buffer followed by centrifugation and careful removal of the supernatant.
- Supernatants are removed and the cell pellets are re-suspended in 100 μl pre-chilled tetramer labeling buffer and placed on ice.
- Solutions of MHC II tetramers loaded with either a validated HIV-1 peptide, or an irrelevant control peptide (CLIP) are spun at full speed for 5 min at 4 °C to remove protein aggregates.
Note: Spinning the MHC II tetramer solution before usage reduces background labeling. - APC-conjugated tetramers are added to the cell suspension at 1 μg per 106 cells (i.e., 5 µg per tube).
- Important: One tube containing 5 x 106 cells has to be labeled with tetramers loaded with an irrelevant peptide (CLIP) and used as a negative control for tetramer labeling.
- Tetramer labeling is performed at 0-4 °C for 60 min, protected from light.
Note: Staining under cold conditions contributes to reduction of background; however, these conditions are appropriate only for the detection of high-avidity pMHC/TCR interactions. - A mixture of antibodies targeting cell surface markers is added to each tube as follows (v/106 cells): 1 μl CD3 APC-eFluor 780; 0.5 μl CD4 PE-CF594; 0.5 μl CD8 BV 785; 3 μl CD14-VioGreen; 3 μl CD20-VioGreen and 0.5 μl Fixable Viability Dye eFluor 506.
Notes:- Additional antibodies for cell surface phenotyping can be added at this point.
- At this point, tubes containing 0.5 x 106 cells in 50 μl FACS buffer are singly labeled with individual antibodies to be used as compensation controls. For the AmCyan-dump channel (including CD14-VioGreen; CD20-VioGreen and Fixable Viability Dye eFluor 506), a single tube containing 0.5 x 106 cells in 50 μl FACS buffer is labeled with 1.5 μl CD14-VioGreen.
- Additional antibodies for cell surface phenotyping can be added at this point.
- Cells are further incubated with the antibody mixture for 30 min at 4 °C in the dark.
- For flow cytometric analysis
- Labeled cells are washed twice with (pre-chilled) FACS buffer and fixed with 2% PFA diluted in PBS.
Note: Fixation entails complete removal of the supernatants following centrifugation and resuspension in 400 µl of the fixation solution. - All cells in the tube are acquired on a flow cytometer (LSR Fortessa).
- Labeled cells are washed twice with (pre-chilled) FACS buffer and fixed with 2% PFA diluted in PBS.
- For cell sorting
- Labeled cells are washed once with FACS sorting buffer and the cell concentration is adjusted to 10 x 106 cells per ml in FACS sorting buffer.
- Cells are filtered using a Cell-Strainer cap to obtain a single-cell suspension. Subsequently, all cells are acquired on a cell sorter (FACSAria II) installed in a microbiological safety cabinet.
- Labeled cells are washed once with FACS sorting buffer and the cell concentration is adjusted to 10 x 106 cells per ml in FACS sorting buffer.
- Viable tetramer-labeled CD4+ T cells are visualized in the viable CD3+, CD4+, CD14-, CD20-, CD8- lymphocyte gate (see gating strategy, Figure 1A).
- Gates are set according to the negative control, consisting in CLIP-loaded tetramer labeled CD4+ T cells (Figure 1B).
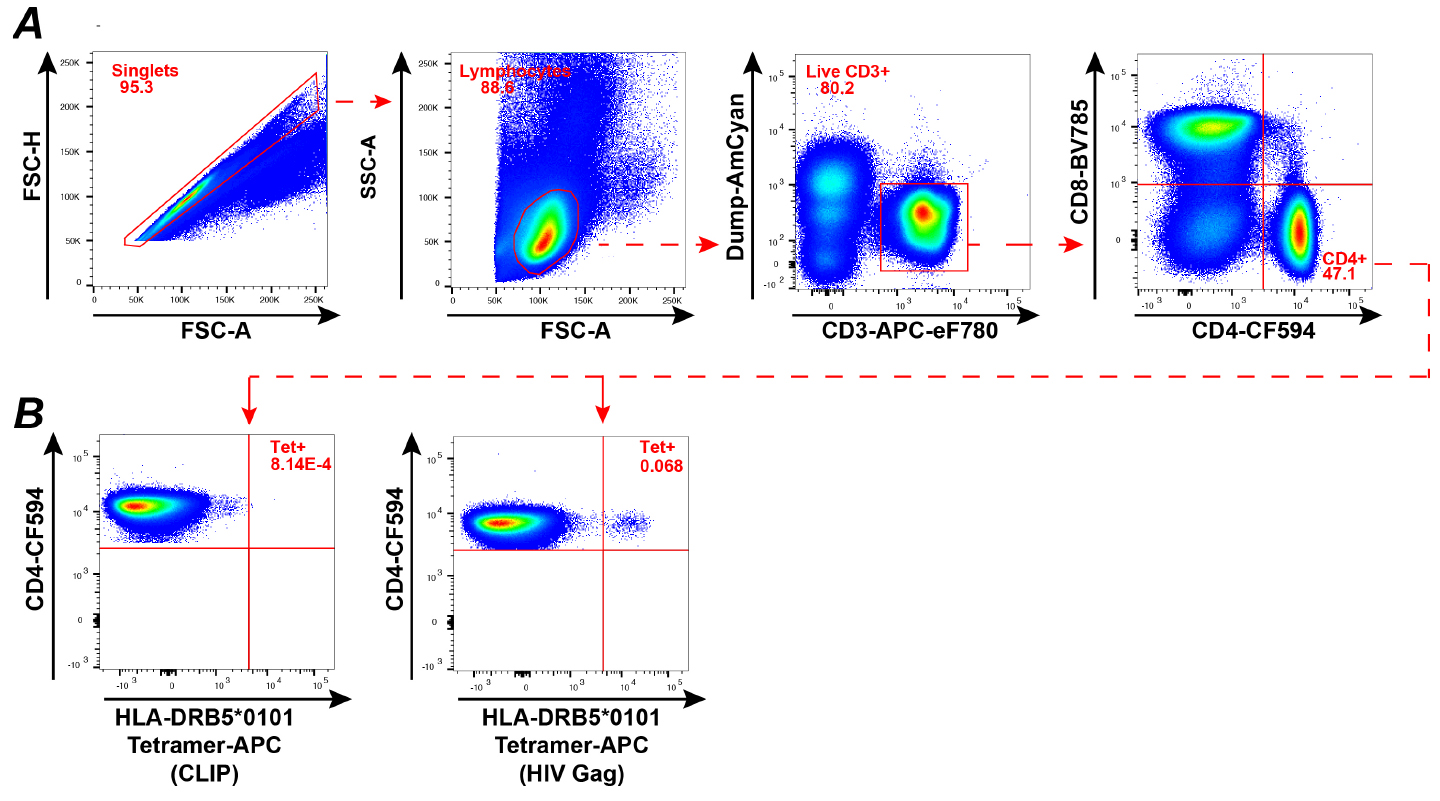
Figure 1. Ex vivo detection of viable HIV-specific CD4+ T cells from patient PBMC. A. Gating strategy: The Dump channel includes the following antibodies: CD14-VioGreen, CD20-VioGreen and viability dye-eFluor 506. B. FACS plots depicting CD4+ T cells from an HIV-1 infected patient labeled with an HLA DRB5*0101 tetramer loaded either with a control CLIP peptide [PVSKMRMATPLLMQA] (left) or with an immunodominant HIV-1 Gag peptide [FRDYVDRFYKTLRAEQASQE] (right).
Data analysis
Flow cytometry data was analyzed with the Flowjo v10.2 software (FlowJo, LLC.).
Recipes
- Complete RPMI
440 ml RPMI 1640
50 ml FBS
5 ml penicillin-streptomycin
5 ml HEPES - FACS buffer (For 1 L)
982 ml 1x PBS
10 g BSA
18 ml 5% (w/v) sodium azide - Tetramer labeling buffer (1 ml)
850 μl RPMI (no FBS)
150 μl human AB serum - FACS sorting buffer (50 ml)
49.5 ml 1x PBS
0.5 ml FBS
Acknowledgments
This protocol was adapted from Benati et al. (2016). We acknowledge funding from Agence Nationale de la Recherche sur le SIDA et les Hépatites Virales (ANRS EP36-8) and Agence Nationale de la Recherche (ANR PD1VAX).
References
- Benati, D., Galperin, M., Lambotte, O., Gras, S., Lim, A., Mukhopadhyay, M., Nouel, A., Campbell, K. A., Lemercier, B., Claireaux, M., Hendou, S., Lechat, P., de Truchis, P., Boufassa, F., Rossjohn, J., Delfraissy, J. F., Arenzana-Seisdedos, F. and Chakrabarti, L. A. (2016). Public T cell receptors confer high-avidity CD4 responses to HIV controllers. J Clin Invest 126(6): 2093-2108.
- Seth, N., Kaufmann, D., Lahey, T., Rosenberg, E. S. and Wucherpfennig K. W. (2005). Expansion and contraction of HIV-specific CD4 T cells with short bursts of viremia, but physical loss of the majority of these cells with sustained viral replication. J Immunol 175(10): 6948-58.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Galperin, M., Benati, D., Claireaux, M., Mukhopadhyay, M. and Chakrabarti, L. A. (2017). MHC Class II Tetramer Labeling of Human Primary CD4+ T Cells from HIV Infected Patients. Bio-protocol 7(6): e2187. DOI: 10.21769/BioProtoc.2187.
Category
Immunology > Immune cell staining > Flow cytometry
Cell Biology > Cell staining > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link