- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of the Predatory Capability of Bdellovibrio bacteriovorus HD100
Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2177 Views: 11614
Reviewed by: Dennis NürnbergAmit DeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Intracellular Sodium Ion Measurement using CoroNa Green
Yusuke V. Morimoto [...] Tohru Minamino
Jan 5, 2017 10438 Views

Determination of Survival of Wildtype and Mutant Escherichia coli in Soil
Yinka Somorin and Conor O'Byrne
Jul 20, 2017 8829 Views

Shipment of Cyanobacteria by Agarose Gel Embedding (SCAGE)—A Novel Method for Simple and Robust Delivery of Cyanobacteria
Phillipp Fink [...] Karl Forchhammer
Dec 5, 2024 1389 Views
Abstract
Bdellovibrio bacteriovorus HD100 is an obligate predator that preys upon a wide variety of Gram negative bacteria. The biphasic growth cycle of Bdellovibrio includes a free-swimming attack phase and an intraperiplasmic growth phase, where the predator replicates its DNA and grows using the prey as a source of nutrients, finally dividing into individual cells (Sockett, 2009). Due to its obligatory predatory lifestyle, manipulation of Bdellovibrio requires two-member culturing techniques using selected prey microorganisms (Lambert et al., 2003). In this protocol, we describe a detailed workflow to grow and quantify B. bacteriovorus HD100 and its predatory ability, to easily carry out these laborious and time-consuming techniques.
Keywords: Bdellovibrio bacteriovorusBackground
In the last years, Bdellovibrio has attracted the interest of the scientific community and several applications have been developed, such as evolution studies (Davidov and Jurkevitch, 2009), identification of new biocatalysts (Martínez et al., 2012), therapeutic applications (Atterbury et al., 2011), or biotechnological applications using Bdellovibrio as a lytic agent for the recovery of value added intracellular bioproducts (Martínez et al., 2016). Due to the growing interest in Bdellovibrio, different indirect methods to quantify this predatory bacterium have been developed (Mahmoud et al., 2007; Lambert and Sockett, 2008; Van Essche et al., 2009). However, direct quantification of Bdellovibrio via double-layer method is still necessary to thoroughly characterize Bdellovibrio predatory capability. Here, we describe a well-established, reliable, and broadly used method that allows Bdellovibrio cell number quantification in predatory co-cultures.
Materials and Reagents
- 0.45 µm sterilization filter (Sartorius, catalog number: 16555-K )
- 0.22 µm sterilization filter (Sartorius, catalog number: 16532-K )
- 10-ml glass test tubes (Fisher Scientific, catalog number: 15175134 )
- Glass microscope slides (76 x 26 mm) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 10143562BEF ) (see Note 1)
- Glass microscope coverslips (22 x 22 mm) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3306 )
- 10 ml syringe (BD, catalog number: 307736 )
- Bdellovibrio bacteriovorus HD100 (ATCC, catalog number: 15356 ) (Stolp and Starr, 1963)
- Pseudomonas putida KT2440 (ATCC, catalog number: 47054 ) (Nelson et al., 2002)
- Glycerol (EMD Millipore, catalog number: 104094 )
- Bacto tryptone (BD, BactoTM, catalog number: 211705 )
- Yeast extract (Conda, catalog number: 1702 )
- Sodium chlorice (NaCl) (EMD Millipore, catalog number: 106404 )
- Sodium hydroxide (NaOH) pellets for analysis (EMD Millipore, catalog number: 106498 )
- Agar (BD, BactoTM, catalog number: 214010 )
- Nutrient broth (NB) (BD, DifcoTM, catalog number: 234000 )
- Calcium chloride dihydrate (CaCl2·2H2O) (EMD Millipore, catalog number: 102382 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (EMD Millipore, catalog number: 105833 )
- HEPES buffer (Sigma-Aldrich, catalog number: H3375 )
- Lysogeny broth (LB) medium (1 L, pH 7.5) (see Recipes)
- LB, 1.5% (w/v) agar (see Recipes)
- NB medium (1 L) (see Recipes)
- CaCl2 and MgCl2 salts (see Recipes)
- Diluted nutrient broth (DNB) medium (1 L, pH 7.4) (see Recipes)
- DNB, 0.7% (w/v) agar (see Recipes)
- DNB, 1.5% (w/v) agar (see Recipes)
- HEPES buffer (see Recipes)
Equipment
- Centrifuge (Eppendorf, model: 5810 R )
- 100 ml flasks
- 30 °C chamber (JP Select, catalog number: 001257 )
- 30 °C shaking incubator (250 rpm) (Eppendorf, New BrunswickTM, model: Innova® 44 )
- Water bath (JP Select, catalog number: 6000138 )
- Phase contrast microscopy (Nikon Instruments, model: OPTIFHOT-2 ) (Note 1)
- Spectrophotometer (Shimadzu, model: UV-260 )
- Leica DFC345 FX camera (Leica Microsystems, model: DFC345 FX )
- Autoclave
Procedure
- Culture of Bdellovibrio preying upon a prey (Figures 1A and 1B)
- Preparation of prey cell suspension (Figure 1A)
- In this example, P. putida KT2440 is used as prey. To obtain cell suspensions, grow the prey in NB medium at 30 °C and 250 rpm for 16 h (see Note 2), centrifuge (30 min, 4000 x g, 4 °C) and resuspend to OD600 = 10 in HEPES buffer (see Note 3).
- Prey cells can be stored at 4 °C for up to 2 weeks. Immediately prior to use, dilute the cells to OD600 = 1.
- Preparation of the preinocule of Bdellovibrio (two-step cultivation) (Figure 1B)
- Recover Bdellovibrio from glycerol stocks stored at -80 °C (see Note 4) by adding 50 µl directly to 10 ml of prey cell suspension prepared in DNB medium at OD600 = 1. Incubate at 30 °C and 250 rpm.
- After 24 h of predation, transfer 100-300 µl of the co-culture to 10 ml of prey suspension prepared in HEPES buffer at OD600 = 1 (predator-prey ratio of 1:10). Incubate at 30 °C and 250 rpm for 24 h.
Note: Set up the co-cultures by adding 10 ml of suspension to 100 ml flasks. - Isolation of Bdellovibrio cells (Figure 1B). After predation, filter co-cultures twice through a 0.45 µm filter to recover B. bacteriovorus.
Note: Use a new filter for the second filtration step. - Set up the co-cultures of interest: transfer 100-300 µl of Bdellovibrio cells to 10 ml of prey suspension prepared in HEPES buffer at OD600 = 1 (predator-prey ratio of 1:10). Incubate at 30 °C and 250 rpm for 24 h.
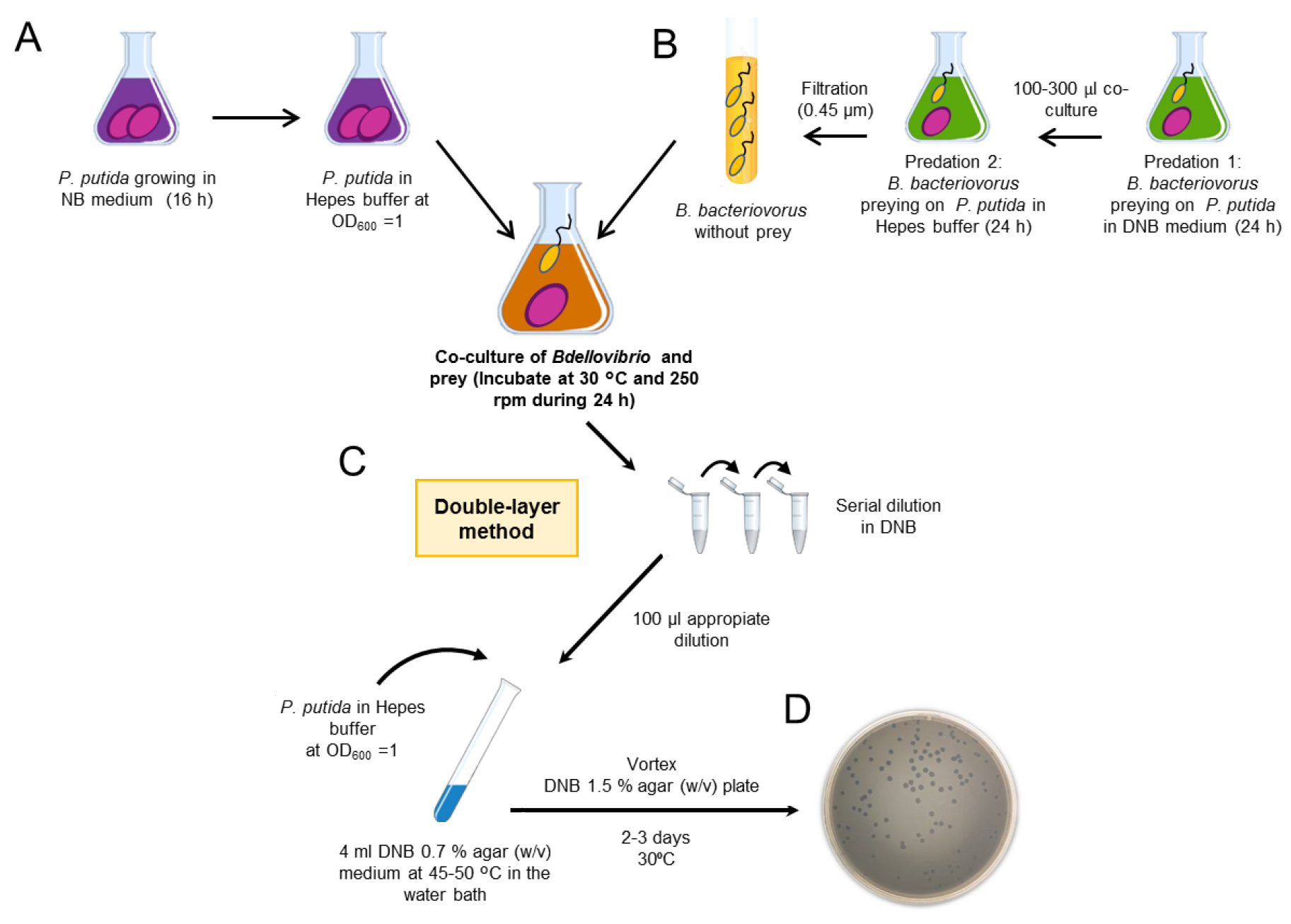
Figure 1. Workflow to set up Bdellovibrio co-cultures. A. Preparation of the prey cell suspension (here, P. putida KT2440). B. Preparation of Bdellovibrio cells. Two-step cultivation of Bdellovibrio is needed to obtain the predatory cells for the experiment. C. Double layer method to quantify the cell number of B. bacteriovorus HD100. D. Development of B. bacteriovorus HD100 on a lawn of prey on DNB agar plates after 2-3 days of incubation at 30 °C. Bdellovibrio cell number can be quantified as plaque-forming-units (pfu/ml).
- Calculation of the viability of Bdellovibrio and prey cells (Figure 1C)
Make serial dilutions of the co-cultures from 10-1 to 10-7 (see Note 5) in DNB medium.- Determine Bdellovibrio viability using the double layer method (see Figure 1C) (Lambert et al., 2003):
- Add 4 ml of DNB 0.7% (w/v) agar into a glass tube and keep at 45-50 °C in a water bath (see Note 6).
- Add 0.5 ml of prey cell suspension (P. putida KT2440 prepared at OD600 = 10 in HEPES buffer; see Figure 1A).
- Add 0.1 ml of the appropriate dilution from the co-cultures.
- Vortex the tube gently.
- Rapidly, pour the mixture onto a DNB 1.5 % (w/v) agar plate.
- Incubate the plates for 2-3 days at 30 °C. Bdellovibrio growth is monitored as plaque-forming units per milliliter (pfu/ml) developing on a lawn of the prey (Figure 1D).
- Calculation of prey cell viability
Place 10 µl of each dilution on LB agar plates (see Note 5) and incubate them at 30 °C. Prey cells are counted as colony-forming units per milliliter (cfu/ml).
Data analysis
A representative graph of the predatory activity of Bdellovibrio is shown below (Figure 2). Predator and prey cell number at the beginning of the experiment (0 h) and after 24 h of predation upon P. putida KT2440 are represented.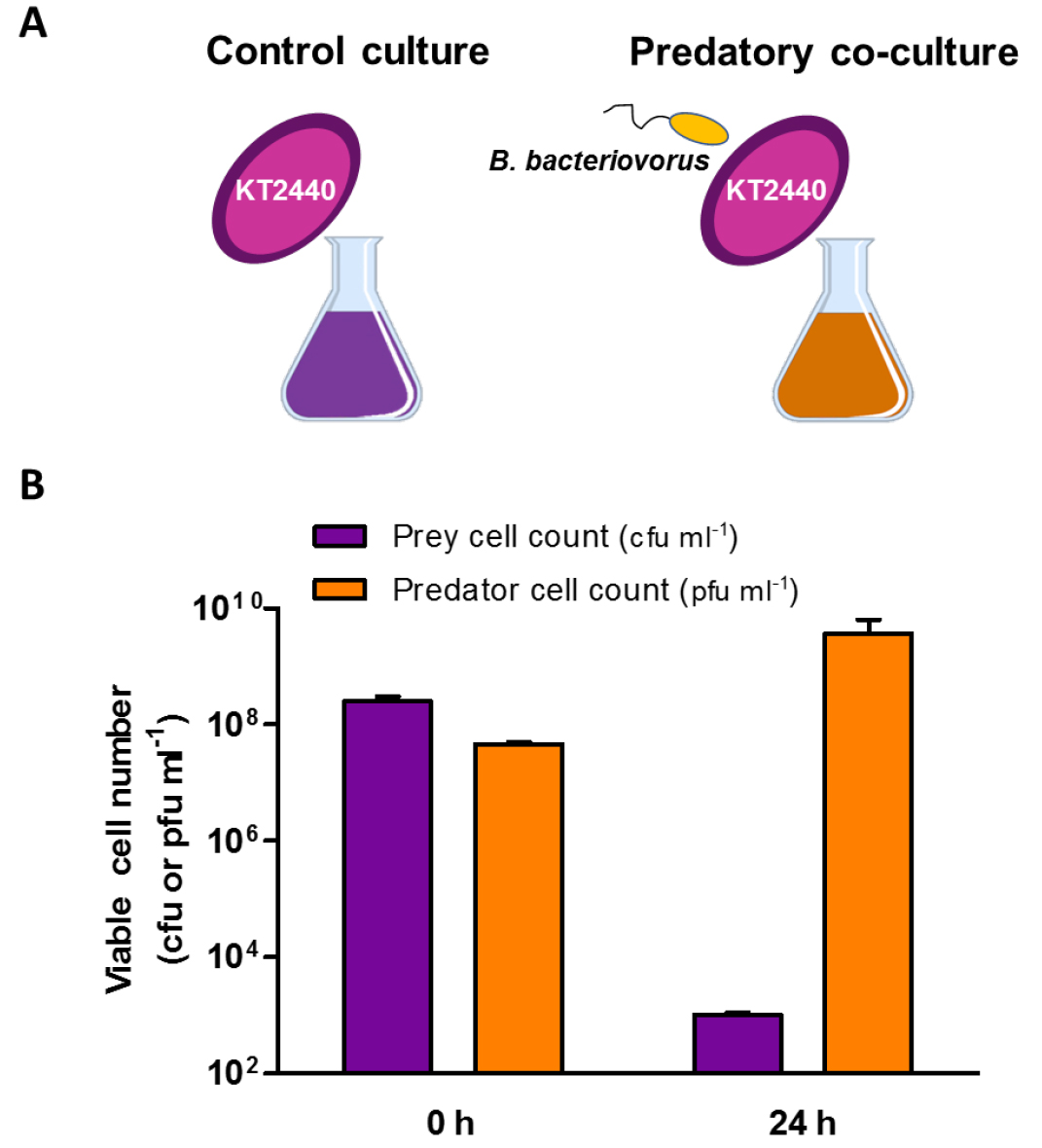
Figure 2. Predation profile to study the predatory capability of B. bacteriovorus HD100 upon P. putida KT2440. A. Schematic representation of the co-culture involving B. bacteriovorus HD100 and the control culture of P. putida without predator. B. Viability of the prey and predator cells at the beginning of the experiment (0 h) and after 24 h of incubation. The purple bars represent the viability of the prey cells and the orange bars correspond to the predator viable cell number. Error bars indicate the standard deviation of the mean (n ≥ 3).
Notes
- If possible, predation events should be checked using a phase contrast microscope to ensure the success of the following steps (see Figure 3).
- Select the culture medium based on the prey cells used to grow Bdellovibrio.
- To measure OD600, prepare a 1/10 dilution of the culture of interest. E.g., Add 100 µl of the culture to 900 µl of saline solution and mix.
- To store Bdellovibrio at -80 °C, add 0.3 ml of 85 % (w/v) glycerol and 0.7 ml of the co-culture of Bdellovibrio and the prey bacteria to a cryogenic vial, and place the tube directly in the -80 °C freezer. Bdellovibrio cells can be revived by simply scratching the ice of the cryogenic vial with a sterile loop and adding it to the prey suspension. There is no need to de-freeze the cryogenic vial for the inoculation.
- Dilute samples further if a high concentration of cells is expected. E.g., Prepare 1:10 serial dilutions by adding 100 µl of the co-culture to 900 µl of DNB medium in Eppendorf tubes and repeat until the desired concentration is obtained.
- DNB 0.7 % (w/v) agar tubes can be previously prepared and stored at room temperature. Prior to use, melt the tubes in a water bath at 150-200 °C and keep them at 45-50 °C.
- Prepare DNB medium (or HEPES buffer) and the CaCl2 and MgCl2 salts separately. Add the salts immediately prior to use.
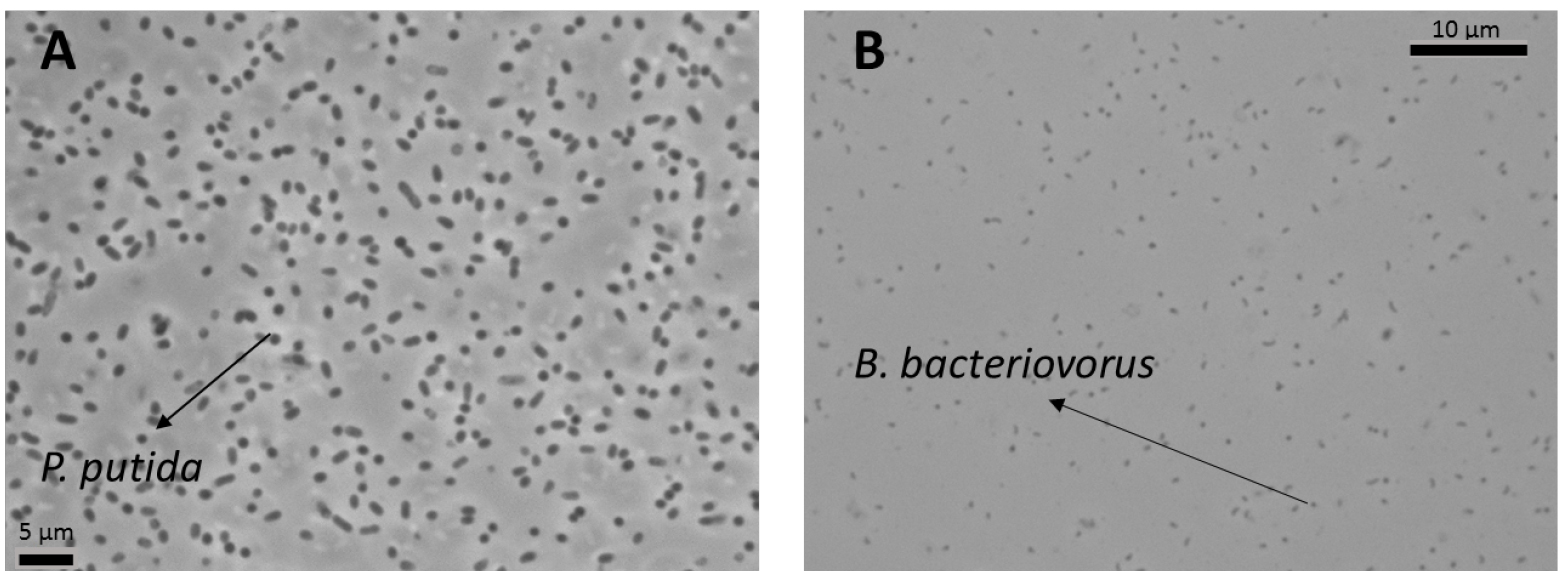
Figure 3. Co-culture of B. bacteriovorus HD100 preying on P. putida KT2440 under the microscope. A. Co-culture at the onset of predation (time zero); B. After 24 h of incubation, only predatory cells can be observed. Cultures are routinely visualized using a 100x phase-contrast objective and images taken with a Leica DFC345 FX camera.
Recipes
- Lysogeny broth (LB) medium (1 L)
10 g Bacto tryptone
5 g yeast extract
10 g NaCl
Adjust to pH 7.5 using 1 N NaOH
Autoclave at 121 °C for 20 min - LB, 1.5% (w/v) agar (1 L)
10 g Bacto tryptone
5 g yeast extract
10 g NaCl
Adjust to pH 7.5 using 1 N NaOH
15 g agar
Autoclave at 121 °C for 21 min - NB medium (1 L)
8 g NB
Autoclave at 121 °C for 20 min - CaCl2 and MgCl2 salts
2 mM CaCl2·2H2O
3 mM MgCl2·3H2O
Sterilize by filtration through a 0.22 µm filter into a sterile container - Diluted nutrient broth (DNB) medium (1 L) (see Note 7)
0.8 g NB
Adjust to pH 7.4 using 1 N NaOH
Autoclave
Add CaCl2 and MgCl2 salts (see Recipe 4) - DNB, 0.7% (w/v) agar (1 L) (see Note 7)
0.8 g NB
Adjust to pH 7.4 using 1 N NaOH
7 g agar
Autoclave at 121 °C for 20 min
Add CaCl2 and MgCl2 salts (see Recipe 4) - DNB, 1.5% (w/v) agar (1 L) (see Note 7)
0.8 g NB
Adjust to pH 7.4 using 1 N NaOH
15 g agar
Autoclave at 121 °C for 20 min
Add CaCl2 and MgCl2 salts (see Recipe 4) - HEPES buffer (see Note 7)
25 mM HEPES buffer
Adjust to pH 7.8 using NaOH pellets
Autoclave at 121 °C for 20 min
Add CaCl2 and MgCl2 salts (see Recipe 4)
Acknowledgments
This protocol was modified and adapted from a predatory assay previously described (Martínez et al., 2016; Lambert et al., 2003). This work was funded by the EU Seventh Framework Programme under grant agreement No. 311815 (SYNPOL), and by grants from the Comunidad de Madrid (P2013/MIT2807) and the Spanish Ministerio de Economía y Competitividad, (BIO2010-21049, BIO2013-44878-R).
References
- Atterbury, R. J., Hobley, L., Till, R., Lambert, C., Capeness, M. J., Lerner, T. R., Fenton, A. K., Barrow, P. and Sockett, R. E. (2011). Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol 77(16): 5794-5803.
- Davidov, Y. and Jurkevitch, E. (2009). Predation between prokaryotes and the origin of eukaryotes. Bioessays 31(7): 748-757.
- Gophna, U., Charlebois, R. L. and Doolittle, W. F. (2006). Ancient lateral gene transfer in the evolution of Bdellovibrio bacteriovorus. Trends Microbiol 14(2): 64-69.
- Jurkevitch, E. and Davidov, Y. (2007). Phylogenetic diversity and evolution of predatory prokaryotes. In: Jurkevitch, E. (Ed.). Microbiol Monogr Predatory Prokaryotes Heidelberg. Springer, pp: 11-56.
- Jurkevitch, E., Minz, D., Ramati, B. and Barel, G. (2000). Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl Environ Microbiol 66(6): 2365-2371.
- Lambert, C., Smith, M. C. and Sockett, R. E. (2003). A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol 5(2): 127-132.
- Lambert, C. and Sockett, R. E. (2008). Laboratory maintenance of Bdellovibrio. Curr Protoc Microbiol 9:7B.2.1-7B.2.13.
- Mahmoud, K. K., McNeely, D., Elwood, C. and Koval, S. F. (2007). Design and performance of a 16S rRNA-targeted oligonucleotide probe for detection of members of the genus Bdellovibrio by fluorescence in situ hybridization. Appl Environ Microbiol 73(22):7488.
- Martínez, V., de la Peña, F., García-Hidalgo, J., Mata, I., García, J. L., and Prieto, M. A. (2012). Identification and biochemical evidence of a medium-chain-length polyhydroxyalkanoate depolymerase in the Bdellovibrio bacteriovorus predatory hydrolytic arsenal. Appl Environ Microbiol 78: 6017-6026.
- Martínez, V., Herencias, C., Jurkevitch, E. and Prieto, M. A. (2016). Engineering a predatory bacterium as a proficient killer agent for intracellular bio-products recovery: the case of the polyhydroxyalkanoates. Sci Rep 6: 24381.
- Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A., Fouts, D. E., Gill, S. R., Pop, M., Holmes, M., Brinkac, L., Beanan, M., DeBoy, R. T., Daugherty, S., Kolonay, J., Madupu, R., Nelson, W., White, O., Peterson, J., Khouri, H., Hance, I., Chris Lee, P., Holtzapple, E., Scanlan, D., Tran, K., Moazzez, A., Utterback, T., Rizzo, M., Lee, K., Kosack, D., Moestl, D., Wedler, H., Lauber, J., Stjepandic, D., Hoheisel, J., Straetz, M., Heim, S., Kiewitz, C., Eisen, J. A., Timmis, K. N., Dusterhoft, A., Tummler, B. and Fraser, C. M. (2002). Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4(12): 799-808.
- Sockett, R. E. (2009). Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol 63: 523-539.
- Stolp, H. and Starr, M. P. (1963). Bdellovibrio Bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29: 217-248.
- Van Essche, M., Sliepen, I., Loozen, G., Van Eldere, J., Quirynen, M., Davidov, Y., Jurkevitch, E., Boon, N and Teughels, T. (2009). Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ Microbiol Rep 1(4):228-33.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Herencias, C., Prieto, M. A. and Martínez, V. (2017). Determination of the Predatory Capability of Bdellovibrio bacteriovorus HD100. Bio-protocol 7(6): e2177. DOI: 10.21769/BioProtoc.2177.
Category
Microbiology > Microbial cell biology > Cell viability
Cell Biology > Cell viability > Predation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









