- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In situ Hybridization (ISH) and Quantum Dots (QD) of miRNAs
Published: Vol 7, Iss 4, Feb 20, 2017 DOI: 10.21769/BioProtoc.2138 Views: 11629
Reviewed by: HongLok LungChiara AmbrogioAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Polysome Fractionation to Analyze mRNA Distribution Profiles
Amaresh C. Panda [...] Myriam Gorospe
Feb 5, 2017 31791 Views

miRNA Characterization from the Extracellular Vesicles
Sajni Josson [...] Leland W.K. Chung
Feb 20, 2017 9018 Views

Detection of Individual RNA in Fixed Cells and Tissues by Chromogenic ISH
Meng Jiang [...] Rongqin Ke
Feb 5, 2020 5185 Views
Abstract
miRNA are short non-coding RNA which inhibit translation of mRNA. miRNA regulate several cellular processes. Certain miRNA are known to induce oncogenesis. miRNA can be measured by real-time PCR and be imaged using a combination of in situ hybridization (ISH) and quantum dots (QD). The advantage of using quantum dots is that several miRNA can be simultaneously measured using multiplexed QD. Additionally, miRNA can be visualized in different regions of the tissue. Since miRNA are biomarkers of various disease states, miRNA can be visualized and quantitated in tissue sections for diagnostic and prognostic purposes. Here we describe ISH-QD analysis of tissue sections. Tissue sections from xenografts or clinical specimens are used. These are deparaffinized, treated with Proteinase K and hybridized with a biotin-probe to specific to the miRNA. The in situ hybridization is performed by labeling the biotin-probes and followed by labeling with streptavidin tagged quantum dots. Image acquisition of the quantum dots is performed and analyzed for the miRNA expression levels. Combining ISH and QD gives a powerful tool to detect miRNA in different cells of the tissue.
Keywords: miRNABackground
miRNAs can be easily detected by quantitative real-time PCR or Northern blotting. However, imaging miRNAs has been challenging. Recent advances in quantum dots imaging have made it possible to determine expression of miRNAs in tissues. Using this process, miRNAs can be visualized in different compartments of a tissue, such as tumor, stroma, immune cells, etc. Additionally, miRNAs can be multiplexed to determine co-localization of miRNAs which mediate specific processes, in different cellular regions. Different tissues can be used for ISH-QD such as tissues from xenografts or human clinical samples. Tissues from animal studies are formalin fixed and paraffin embedded. These tissues were used for ISH-QD analysis (Figure 1). 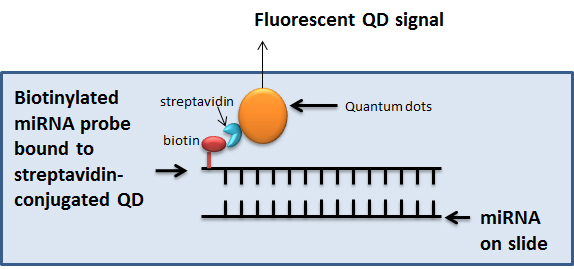
Figure 1. In situ hybridization coupled to Quantum dots labelling for visualization of miRNA. In the ISH-QD protocol, miRNA are detected on formalin fixed paraffin embedded tissue sections using biotinylated miRNA probes binding to streptavidin-conjugated QDs. The specific QD gives a specific fluorescent signal which is quantified using the Inform v1.3 software.
Materials and Reagents
- Gloves
- Tissue slides
- Superfrost plus slides
- Coverslips
- LNATM Scramble-miR probe (Exiqon, catalog number: 699004-370 )
- LNATM miR-409-3p 5’ biotin labeled (Exiqon, catalog number: 610701-370 )
- LNATM miR-409-5p 5’ biotin labeled (Exiqon, catalog number: 615615-370 )
- RNaseZap (Thermo Fisher Scientific, AmbionTM)
- Xylene
- Ethanol
- Sterile PBS (pH 7)
- Nail polish
- Horse serum (Vector Laboratories, catalog number: S-2000 )
- Streptavidin block reagent (Thermo Fisher Scientific, InvitrogenTM)
- Streptavidin conjugated QDs 625 nm (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: A10196 ) (1 µm stock from Invitrogen)
- Streptavidin conjugated QDs 565 nm (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: Q10131MP ) (1 µm stock from Invitrogen)
- 6% IgG-free, protease free BSA (Jackson ImmunoResearch, catalog number: 001-000-162 )
- BSA
- 0.4% Triton X-100
- 0.1% Tween-20
- Mounting media
- 4’6-diamidino-2-phenylindole (DAPI) (Vector Laboratories)
- 1 M Tris-HCl (pH 7.4)
- 0.5 M EDTA
- NaCl
- RNase free Milli-Q water, autoclaved (all solutions prepared in this water)
- Exiqon microRNA ISH buffer set and Proteinase K (Exiqon, catalog number: 90000 )
- 20x SSC, pH 7.0
- Proteinase K stock (see Recipes)
- Proteinase K buffer (see Recipes)
- Proteinase K solution (15 µg/ml) (see Recipes)
- SSC solutions (see Recipes)
- Hybridization mix (see Recipes)
- Streptavidin blocking solution (see Recipes)
- QD solutions (see Recipes)
- PBS-T (see Recipes)
Equipment
- Autoclave
- Hybridizer (DAKO Statspin Hybridizer)
- Glass cutter
- Slide rack and glass jars
- PAP pen or ImmEdge pen (Vector Laboratories, catalog number: H-4000 )
- Water bath
- CRi multi-spectral camera with built-in Nuance software and inForm software (PerkinElmer, Waltham, MA)
Software
- Nuance v3.1 software
- Inform v1.3 software
- Graphpad Prism software
Procedure
Notes:
- The in situ hybridization procedure has been described in detail in the Exiqon ISH protocol.
- Caution of RNA work
- All steps should take place in a clean and nuclease free environment.
- All surfaces should be cleaned with RNaseZap.
- Wear gloves at all times and use autoclaved glassware and RNase grade MilliQ water which is autoclaved for preparation of all solutions.
- In situ hybridization was performed as mentioned in the Exiqon manual.
- Optimization
- Optimization of Proteinase K treatment required before the start of the actual assay. Proteinase K reagent prepared immediately before use at a concentration of 15 µg/ml.
- Optimization of LNA miRNA probe and scramble miRNA. A probe concentration of 80 nM was optimal for detection of miR-409-3p and miR-409-5p.
- Reagents to be prepared on the day of the experiment
- Proteinase K
- Hybridization mix: Place the appropriate amount of probe in a 2 ml non-stick RNase-free tube. Denature the probe at 90 °C for 4 min. Centrifuge the tubes and spin down. Add 2 ml of 1x miRNA ISH buffer.
- Deparaffinize slides in xylene and ethanol
- Xylene - 5 min, 3 times in a glass jar
- 99.9% ethanol - 10 min, 2 times, in a glass jar
- 99.9% ethanol - 5 min, 1 time, in a glass jar
- 96% ethanol - 10 min, 1 time, in a glass jar
- 96% ethanol - 5 min, 1 time, in a glass jar
- 70% ethanol - 10 min, 1 time, in a glass jar
- 70% ethanol - 5 min, 1 time, in a glass jar
- PBS - 2-5 min, each 1 time, in a glass jar
- Xylene - 5 min, 3 times in a glass jar
- Proteinase K treatment and wash
- Add Proteinase K to Proteinase K buffer (final concentration 15 µg/ml) (add 7.5 µl Proteinase K stock to 10 ml Proteinase K buffer).
- Add sufficient volume of proteinase K solution to cover the slides (instructions in the Exiqon manual).
- Incubate for 10 min, at 37 °C, in a Hybridizer.
- Transfer the slides into a new glass jar containing PBS. Repeat this step twice.
- Add Proteinase K to Proteinase K buffer (final concentration 15 µg/ml) (add 7.5 µl Proteinase K stock to 10 ml Proteinase K buffer).
- Hybridization
- Use PAP pen to mark around the tissue section.
- Using a glass cutter cut the coverslip (autoclaved) to the size of the tumor tissue section.
- Add miRNA detection probe (50 µl, hybridization mix) on the tissue section. Avoid air bubbles.
- If a slide has several serial tissues sections of the same tumor, it can be used to determine several probes (such as miR-409-3p, miR-409-5p and Scramble miRNA). Once the probe is added, place the cut coverslip very carefully without trapping air bubbles.
- Seal the edge of the coverslip to the slide using colorless nail polish. Let it dry.
- Place the slides in the Hybridizer at 55 °C for 1 h.
- Use PAP pen to mark around the tissue section.
- Disassembly and washes
- Carefully remove the coverslips.
- Place the slides into a glass jar with 5x SSC in a water bath at 55 °C.
Note: Use a water bath at 55 °C to do the washes in glass jars. The graded SSC solutions were prepared with autoclaved water. - 5x SSC - 5 min 55 °C
- 1x SSC - 5 min 55 °C
- 1x SSC - 5 min 55 °C
- 0.2x SSC - 5 min 55 °C
- 0.2x SSC - 5 min 55 °C
- 0.2x SSC - 5 min RT
- Incubate the slides in PBS.
- Carefully remove the coverslips.
- Blocking
The slides were incubated in PBS containing 2.5% horse serum and 20% streptavidin block reagent for 1 h at room temperature. - Single QD labeling
- Streptavidin-conjugated QDs (565 nm [green] and 625 nm [red]) were prepared at a 10 nM concertation in PBS-T with 6% IgG-free, protease free BSA.
- The slides were incubated at 37 °C for 60 min.
- Streptavidin-conjugated QDs (565 nm [green] and 625 nm [red]) were prepared at a 10 nM concertation in PBS-T with 6% IgG-free, protease free BSA.
- Washing
The slides were washed 3 times for 5 min each in PBS-T (PBS + 0.4% Triton X-100 + 0.1% Tween-20). - Mounting
Cover the slides with cover glass using aqueous mounting media containing 4’6-diamidino-2-phenylindole (DAPI).
- Image acquisition
The CRi spectral imaging system was used. Multispectral images were taken using the Nuance v3.1 software. For each field of the tissue, serial images were taken from 450 to 800 nm with an interval of 10 nm. This generates a cube a stack of 36 separate images at different wavelengths and has spectral information for every pixel in the image. Images are acquired at 400x magnification (Figure 2).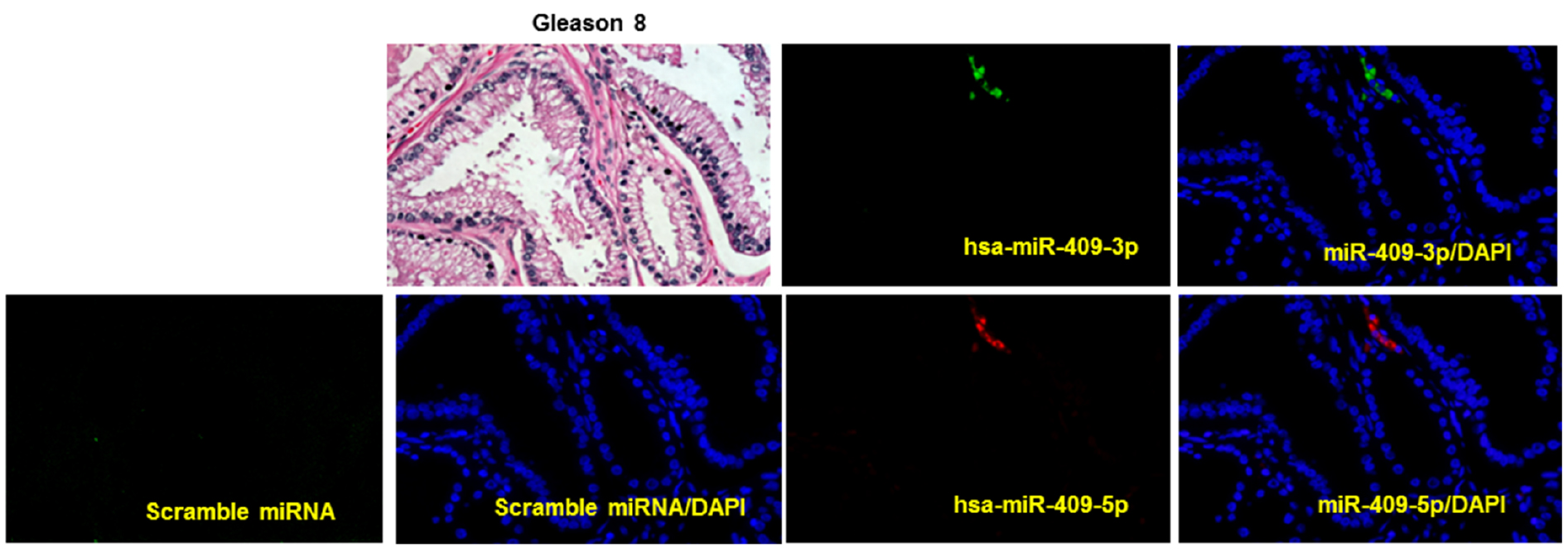
Figure 2. Representative images of multiplexed image of miR-409-3p (green) and miR-409-5p (red) on a human prostate cancer tissue sample of Gleason grade 8, detected using in situ hybridization and quantum dots labeling. DAPI was stained in blue. Scrambled miRNA was used as a negative control, on a adjacent section on an separate slide of the same tissue. Hematoxylin and eosin staining (H&E) was also performed. (Magnification 40x)
Data analysis
Once data acquisition is performed the images are deconvoluted or unmixed to extract the specific signal by a specified QD. A spectral library was built for 565 and 625 nm. The spectral library was used to unmix the cube. Autofluorescence was reduced using the Real Component Analysis plug-in software. The images after unmixing represented the distribution of QDs on the tissue. Signal was quantified using Inform v1.3 software. The software discriminates cancer and stromal (non-cancer) areas. The software also discriminates between nuclear and cytoplasmic area based on the DAPI stain. Using this software the QD signal from the tumor and stromal sections can be determined and quantified. Numerical values are generated for the image and the cytoplasmic component of the tumor and stromal regions is used for plotting the data. The data is graphically plotted using Graphpad Prism software. Data distribution was depicted in box plot formats. Statistical analysis of tissue array was performed using a non-parametric Wilcoxon rank sum test. Values of P < 0.05 were considered to be statistically significant.
Notes
- Multiplexed QD
- Principle: Multiplexed ISH-QD labeling involves using a single tissue which is labeled with a miRNA-biotin probe using ISH and followed by a streptavidin blocking step and incubated with streptavidin conjugated-QD. This process is repeated with another miRNA-biotin probe and another (different wavelength) streptavidin conjugated-QD pair. Finally the tissue is stained with mounting media containing DAPI. Thus, an image is generated for different miRNA with different active fluorescent QD on a single tissue section.
- Procedure: Slides were stained sequentially, with one miRNA probe and a QD procedure followed by another probe and QD. After the first QD labeling, slides were washed 3 times for 5 min each with PBS-T. Slides were then used for hybridization (Procedure C) with another miRNA probe and continued to Procedure H. Hybridization mix and probes stable for only 6 months, avoid multiple freeze-thaw cycles.
- Freshly prepared tissue slides give better miRNA signals. Older tissue preparations, may have loss of miRNA content due to RNase degradation over time.
Recipes
- Proteinase K stock
Reconstitute 20 mg/ml in 600 µl of 10 mM Tris-HCl, pH 7.4 (RNase-free). Store at -20 °C - Proteinase K buffer
5 ml of 1 M Tris-HCl (pH 7.4)
2 ml of 0.5 M EDTA
0.2 ml of 5 M NaCl
900 ml of RNase-free water
Adjust the volume to 1,000 ml and autoclave - Proteinase K solution (15 µg/ml)
Before the start of the experiment add 7.5 µl of Proteinase K stock to 10 ml Proteinase K buffer to make a concentration of 15 µg/ml - SSC solutions
Stock: 20x SSC solution
5x SSC: 1 L = 250 ml 20x SSC + 750 ml water
5x SSC: 1 L = 50 ml 20x SSC + 950 ml water
5x SSC: 1 L = 10 ml 20x SSC + 990 ml water
All solutions to be autoclaved - Hybridization mix (miRNA ISH buffer and LNATM detection probes)
Note: LNATM miRNA detection probes come in 40 µl volume at a 25 µM concentration. - Add 1 ml 2x ISH buffer to 1 ml of RNase free water
- Denature the LNATM miRNA detection probes at 90 °C for 4 min before adding the ISH buffer. Spin down the solution
- For making 80 nM of miRNA probes, add 6.4 µl of denatured probe (25 µM) to 2 ml of 1x ISH buffer
Note: Hybridization mix can be stored at -20 °C for six months. - Streptavidin blocking solution
PBS containing 2.5% horse serum and 20% streptavidin block reagent - QD solutions
Streptavidin-conjugated QDs (565 nm [green] and 625 nm [red]) was prepared at a 10 nM concertation in PBS with 6% IgG-free, protease free BSA - PBS-T (PBS + 0.4% Triton X-100 + 0.1% Tween-20)
Add 1 ml of Tween-20 and 4 ml of Triton X-100 to 1 L PBS
Autoclave at 121 °C for 15 min
Acknowledgments
This procedure has been published in part in the paper by Hu et al., 2011 and Josson et al., 2015. The in situ hybridization procedure has been optimized from the Exiqon ISH protocol. Grant support for this work is from P01-CA98912, DAMD-17-03-02-0033, RO1-CA122602 (L.W.K. Chung).
References
- Hu, P., Chu, G. C., Zhu, G., Yang, H., Luthringer, D., Prins, G., Habib, F., Wang, Y., Wang, R., Chung, L. W. and Zhau, H. E. (2011). Multiplexed quantum dot labeling of activated c-Met signaling in castration-resistant human prostate cancer. PLoS One 6(12): e28670.
- Josson, S., Gururajan, M., Sung, S. Y., Hu, P., Shao, C., Zhau, H. E., Liu, C., Lichterman, J., Duan, P., Li, Q., Rogatko, A., Posadas, E. M., Haga, C. L. and Chung, L. W. (2015). Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 34(21): 2690-2699.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Josson, S., Gururajan, M. and Chung, L. W. (2017). In situ Hybridization (ISH) and Quantum Dots (QD) of miRNAs. Bio-protocol 7(4): e2138. DOI: 10.21769/BioProtoc.2138.
Category
Cancer Biology > General technique > Biochemical assays > RNA
Biochemistry > RNA > miRNA tagging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












